Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 15
Association of Long Non-Coding RNAs (lncRNAs) ANRIL and MALAT1 Polymorphism with Cervical Cancer
Authors Yao Y, Liang Y, Dong X, Liu S, Zhang S, Liu W, Li Y, Shi L, Yan Z, Yao Y
Received 24 January 2022
Accepted for publication 11 April 2022
Published 21 April 2022 Volume 2022:15 Pages 359—375
DOI https://doi.org/10.2147/PGPM.S358453
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Yueting Yao,1,* Yan Liang,2,* Xudong Dong,3 Shuyuan Liu,1 Shao Zhang,4 Weipeng Liu,1 Yu Li,5 Li Shi,1 Zhiling Yan,4 Yufeng Yao1
1Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming, 650118, People’s Republic of China; 2College of Nursing Health Sciences, Yunnan Open University, Kunming, 650223, People’s Republic of China; 3The First People’s Hospital of Yunnan Province & The Affiliated Hospital of Kunming University of Science and Technology, Kunming, 650032, People’s Republic of China; 4Department of Gynaecologic Oncology, The Third Affiliated Hospital of Kunming Medical University, Kunming, 650118, People’s Republic of China; 5Department of Obstetrics, The First People’s Hospital of Kunming, Kunming, 650011, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhiling Yan, Department of Gynaecologic Oncology, The Third Affiliated Hospital of Kunming Medical University, Kunming, Yunnan, 650118, People’s Republic of China, Email [email protected] Yufeng Yao, Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming, Yunnan, 650118, People’s Republic of China, Email [email protected]
Background: Long non-coding RNAs (lncRNAs) and their polymorphisms play crucial roles in the development of different cancers.
Methods: Eight single-nucleotide polymorphisms (SNPs) in ANRIL and MALAT1 (rs1333045, rs4977574, rs1333048, and rs10757278 in ANRIL and rs11227209, rs619586, rs664589, and rs3200401 in MALAT1) were enrolled and genotyped in a total of 1248 samples, including 587 patients with cervical cancer (CC) and 661 healthy individuals using in TaqMan assay. The association of these SNPs with CC was then evaluated.
Results: Our results showed that the allele and genotype frequencies of rs3200401 in MALAT1 were significantly different between the control and CC groups after Bonferroni correction (P = 0.001 and P = 0.004, respectively), indicating that the C allele is a protective factor against CC (OR = 0.70; 95% CI = 0.57– 0.87). In addition, the allele and genotype frequencies of rs4977574 in ANRIL were significantly different between the control and CC groups after Bonferroni correction (P = 0.004 and P = 0.014, respectively), and the A allele might be a protective factor for CC (OR = 0.80; 95% CI = 0.68– 0.93). For subgroup analysis, the alleles of rs3200401 in MALAT1 showed significant differences between the control and adenocarcinoma (AC) and control and squamous cell carcinoma (SCC) groups (P = 0.005 and P = 0.004, respectively). The rs3200401C allele could be a protective factor for AC and SCC development (OR = 0.57; 95% CI = 0.38– 0.85; OR = 0.72; 95% CI = 0.58– 0.90). Moreover, the rs3200401C allele could be a protective factor for cervical cancer stage I development (OR = 0.67; 95% CI = 0.53– 0.86).
Conclusion: Our results indicate that rs3200401 in MALAT1 and rs4977574 in ANRIL could play key roles in the CC development.
Keywords: cervical cancer, single-nucleotide polymorphisms, long non-coding RNAs, association study
Introduction
Cervical cancer (CC) is the fourth common cancer among women. China has contributed to an increasing global cervical cancer burden, with 106,000 cases and 48,000 deaths.1 Cervical cancer includes two main pathological types: squamous cell carcinoma (SCC) and adenocarcinoma (AC). SCC is the most common pathological type which covers 75% to 80% of cases, followed by AC, which accounts for 10.0% to 25.0%.2
Recently, a genome-wide association study found that host genetic factors could play an important role in the CC development.3–7 Many studies have focused on the dysregulation of long non-coding RNAs (lncRNAs) in host genes, because they play critical regulatory roles in cancers as either tumour suppressors or oncogenic lncRNAs.8,9 lncRNA antisense non-coding RNA in the INK4 locus (ANRIL) is located in the chromosome 9p21, which is associated with different cancers.10–15 Besides the ANRIL, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), located on chromosome 11q13.1, participates in epigenetic changes, changes in gene expression, and alternative splicing, which is also associated with different cancers.16,17
Many studies have reported that the polymorphisms of lncRNAs could influence the interaction between lncRNAs and other molecules and are associated with cancer development.18–20 In 2017, Taheri et al investigated the association between four ANRIL SNPs (rs1333045, rs4977574, rs1333048, and rs10757278) and benign prostate hyperplasia and prostate cancer.21 Their results showed that rs4977574, rs1333048, and rs10757278 were associated with benign prostate hyperplasia and prostate cancer risk.21 In 2019, Qu et al investigated the association between MALAT1 tagSNPs (rs11227209, rs619586, rs664589, and rs3200401) and oesophageal squamous cell carcinoma (ESCC).22 Their results showed that rs3200401C is associated with an increased risk of ESCC.22
In the current study, we genotyped four SNPs (rs1333045, rs4977574, rs1333048, and rs10757278) in ANRIL and four tagSNPs (rs11227209, rs619586, rs664589, and rs3200401) in MALAT1 in patients with CC and healthy individuals. We then analyzed the association of these eight SNPs with CC development.
Materials and Methods
Ethics Declarations
This study was approved by the Institutional Review Board of the Third Affiliated Hospital of Kunming Medical University. The study protocol was in accordance with the principles of the Helsinki Declaration of 1975, as revised in 2008.
Subjects
A total of 1248 subjects, including 661 healthy control and 587 patients with CC were recruited for this study. All patients were diagnosed at the Third Affiliated Hospital of Kunming Medical University between 2013 and 2018. The diagnosis of CC was based on the World Health Organization Comprehensive Cervical Cancer Control: A Guide to Essential Practice and the International Federation of Gynaecology and Obstetrics. The inclusion criteria for healthy control was as follows: (1) lack of cancer history and lesion; (2) HPV negative. All subjects were Han Chinese from Yunnan Province, and all signed informed consent forms.
DNA Extraction
Venous blood (5 mL) was collected from each subject, all of whom had fasted. Genomic DNA from peripheral blood was extracted using genomic DNA mini kit (QIAamp DNA Blood Mini Kit). An ultramicro UV-visible spectrophotometer (ND-2000, Thermo Scientific, USA) was used to detect the concentration and purity of the DNA.
SNP Genotyping
The probes and primers used for genotyping these eight SNPs were all purchased from ABI (http://www.appliedbiosystems.com). Primers and probes for the genotyping were commercially available. The array ID were C__32062431_10 (rs11227209), C__1060479_10 (rs619586), C__1060482_20 (rs664589), C__3246069_10 (rs3200401), C__1754667_10 (rs1333048), C__1754681_10 (rs4977574), C__8766826_10 (rs1333045), C__11841860_10 (rs10757278). The TaqMan Genotyping Master Mix used in the genotyping was purchased from ABI, and the genotyping process was the same as that used in our previous study.23 The PCR experiment data were analyzed using TaqMan Genotyper Software (Version 1.3.1). Several samples of each genotype were sequenced to identify the accuracy of SNP genotyping. Each genotype sample was then used as a positive sample in TaqMan assay.
Statistical Analysis
Differences in age between the CC and control groups were analyzed using the Student’s t-test. Hardy-Weinberg equilibrium (HWE) was calculated using SHEsis program.24,25 The associations of eight SNPs with CC were calculated by SHEsis program.24,25 The inheritance model analysis is an attempt to identify the mode of inheritance of association between eight SNPs genotypes and CC which was used by SNPstats software.26 The inheritance models included codominant, dominant, recessive, overdominant, and log-additive models. The best fit model for each SNP was evaluated using the Akaike information criterion (AIC) and the Bayesian information criterion (BIC), and the smallest AIC and BIC were the best fit models.26 Bonferroni correction was applied for multiple comparisons. The differences were statistically significant at P<0.006 (0.05/8).
Results
Subject Characteristics
Table 1 shows the clinical data of the participants in this study. The CC group contained 587 patients, including 488 with squamous cell carcinoma (SCC), 76 with adenocarcinoma (AC), and 23 with other types of cancer. According to CC staging, 349 patients were in stage I, 206 in stage II, and 32 in stage III+IV. The average ages for CC and control groups were 46.12±9.34 and 46.80±6.91 respectively. No significant difference in age was found between the two groups (P=0.153).
 |
Table 1 Characteristics of the Subjects Enrolled in the Current Study |
Association Analysis of Eight SNPs in the MALAT1 and ANRIL in Control and CC Groups
The alleles and genotype frequencies of the eight SNPs in MALAT1 and ANRIL between the control and CC groups are shown in Table 2. The results of HWE show that the four SNPs in MALAT1 and four SNPs in ANRIL were all in HWE in both the control and CC groups (P>0.05) (Table 2). For MALAT1, the allele and genotype frequencies of rs3200401 were significantly different between the control and CC groups after Bonferroni correction (P=0.001 and P=0.004, respectively), and the C allele might be a protective factor against CC (OR=0.70; 95% CI=0.57–0.87). For ANRIL, the allele and genotype frequencies of rs4977574 were significantly different between the control and CC groups after Bonferroni correction (P=0.004 and P=0.014, respectively), and the A allele might be a protective factor for CC (OR=0.80; 95% CI=0.68–0.93). For the other six SNPs, there were no difference in alleles and genotypes between these two groups after Bonferroni correction (P>0.006).
 |
Table 2 The Allele and Genotype Distribution of the Eight SNPs in Control and Cervical Cancer Groups |
Inheritance Model Analysis of Eight SNPs in the MALAT1 and ANRIL in Control and CC Groups
The inheritance models were used to analyze the association of these SNP genotypes with CC. The results of the inheritance model analysis of eight SNPs in MALAT1 and ANRIL between the control and CC groups are shown in Tables 3 and 4. Comparison between CC and control groups showed that the log-additive model was the best fit model for rs3200401 in MALAT1. In this model, the 2T/T+C/T genotype was associated with an increased CC risk (P=0.001, OR=1.42; 95% CI=1.15–1.75). The best fit for rs4977574 in ANRIL was the log-additive inheritance model. In this model, the 2G/G+A/G genotype was associated with an increased CC risk (P=0.003, OR=1.28; 95% CI=1.09–1.50). There were no significant differences of other SNPs between the control and CC groups in the inheritance models (P>0.006).
 |
Table 3 The Inheritance Model Analysis for the Four SNPs in MALAT1 Between CC and Control Groups |
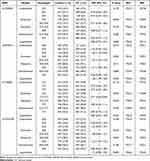 |
Table 4 The Inheritance Model Analysis for the Four SNPs in ANRIL Between CC and Control Groups |
Association Analysis of Eight SNPs in the MALAT1 and ANRIL with Different CC Pathological Types
A comparison between patients with different CC pathological types is shown in Tables 5 and 6. However, the alleles and genotypes of rs3200401 in MALAT1 were significantly different between the control and AC groups (P=0.005 and P=0.005, respectively). The rs3200401C allele could be a protective factor for AC development (OR=0.57; 95% CI=0.38–0.85). In addition, the alleles of rs3200401 in MALAT1 were significantly different between the control and SCC groups (P=0.004). The rs3200401C allele could be a protective factor for SCC development (OR=0.72; 95% CI=0.58–0.90). The allelic frequency and genotype frequency of eight SNPs in MALAT1 and ANRIL were not significantly different between the SCC and AC groups (P>0.006).
 |
Table 5 The Allele and Genotype Distribution of the Eight SNPs in Control and SCC Groups |
 |
Table 6 The Allele and Genotype Distribution of the Eight SNPs in Control and AC Groups |
Inheritance Model Analysis of Eight SNPs in the MALAT1 and ANRIL with Different CC Pathological Types
The results of the analysis of different pathological types of cervical cancer under different inheritance models are shown in Tables 7 and 8. Comparison between the control and SCC groups showed that the log-additive model was the best fit model for rs3200401 in MALAT1. In this model, the 2T/T+C/T genotype was associated with an increased risk of cervical cancer (P=0.004, OR=1.38; 95% CI=1.11–1.73). There were no significant differences between the AC and control groups, and between AC and SCC groups of the eight SNPs in the inheritance model (P>0.006).
 |
Table 7 The Inheritance Model Analysis Between Control and SCC Groups |
 |
Table 8 The Inheritance Model Analysis Between Control and AC Groups |
Association Analysis of Eight SNPs in the MALAT1 and ANRIL with Different Stages of Cervical Cancer
As only 32 patients had stage III + IV cervical cancer, we analyzed the association of the eight SNPs in MALAT1 and ANRIL for patients with stage I and stage II cervical cancer. The alleles and genotypes of rs3200401 in MALAT1 were significantly different between the control and stage I groups (P=0.001 and P=0.005, respectively) (Table 9). The rs3200401C allele could be a protective factor for stage I development (OR=0.67; 95% CI=0.53–0.86) (Table 9). The allelic and genotypic frequencies of the eight SNPs were not significantly different between the control and stage II, and stage I and stage II (P>0.006) (data not shown).
 |
Table 9 The Allele and Genotype Distribution of the SNPs in Control and CC Stage I Groups |
Inheritance Model Analysis of Eight SNPs in the MALAT1 and ANRIL with Different Stages of Cervical Cancer
Comparison between the control and stage I groups showed that the log-additive model was the best fit model for rs3200401 in MALAT1 (Table 10). In this model, the 2T/T+C/T genotype was associated with an increased CC (P=0.002, OR=1.47; 95% CI=1.16–1.88) (Table 10). There were no significant differences between the control and stage II, and between stage I and stage II of the eight SNPs in the inheritance model (P>0.006) (data not shown).
 |
Table 10 The Inheritance Model Analysis Between Control and CC Stage I Groups |
Discussion
Long non-coding RNAs (lncRNAs) and their polymorphisms play key roles in the development of different cancers. In the current study, we genotyped SNPs in ANRIL and MALAT1 in cervical cancer patients and healthy control individuals. Our results showed that rs3200401 in MALAT1 and rs4977574 in ANRIL could be associated with the development of CC.
Recently, ANRIL was shown to be upregulated in tumour tissue and function as a tumour-promoting lncRNA in different cancers. In 2018, Zhang et al reported that the expression of lncRNA ANRIL was upregulated in cervical cancer tissues and cell lines.15 In addition, the downregulation of ANRIL could suppress the proliferation and invasion of CC cell, and enhance the apoptosis, indicating that ANRIL plays an oncogenic role in CC development.15 In 2017, Khorshidi et al reported that rs4977574 in ANRIL was not associated with breast cancer in Iran.27 However, Taheri et al found that rs4977574 was associated with benign prostate hyperplasia and prostate cancer risk, and the A allele could be a protective factor for benign prostate hyperplasia (P=0.017, OR=0.66; 95% CI=0.47–0.93) and prostate cancer risk (P=0.001, OR=0.58; 95% CI=0.41–0.81).21 In 2018, Huang et al performed a meta-analysis of the association between ANRIL polymorphisms and cancer risk, and their results found that GG (GG vs AA) and the G allele were associated with an increased risk of cancer (P<0.001, OR=2.40; 95% CI =1.60–3.59, P<0.001, OR = 1.68; 95% CI = 1.35–2.08).28 In the current study, we also found that the allele and genotype frequencies of rs4977574 were significantly different between the CC and control groups (P=0.004 and P=0.014, respectively) and that the A allele might be a protective factor for CC (OR=0.80; 95% CI=0.68–0.93). The rs4977574 SNP could alter the binding sites for ETS transcription factors using the HaploReg v4 software by Ward and Kellis.20 The overexpressed ETS family transcription factor ETS-2 was found to be associated with the development of cervical cell neoplasia.29 These results indicated that rs4977574 could influence the interaction between ANRIL and ETS-2, which is associated with CC.
MALAT1 regulates alternative splicing and transcriptional regulation by modulating the levels of active serine/arginine (SR) proteins such as serine/arginine-rich splicing factor 1 (SRSF1) and SRSF2.30,31 MALAT1 is co-localised with SRSF2 splicing domains and is associated with phosphorylation of SRSF2, interacting with SR proteins as a “molecular sponge,” influencing their stability; MALAT1 regulates the alternative splicing of pre-mRNAs.30,31 In 2019, Qu et al investigated the association between MALAT1 tagSNPs (rs11227209, rs619586, rs664589, and rs3200401) and oesophageal squamous cell carcinoma (ESCC), and their results showed that rs3200401C is associated with an increased risk of ESCC.22 In the current study, we also found that the allele and genotype frequencies of rs3200401 in MALAT1 were significantly different between the control and CC groups, which indicates that the C allele is a protective factor against CC. Our results are similar to those of Qu et al rs3200401 is located in MALAT1 M region (6008-7011 nucleotides), which is one of the binding sites of SRSF2.18 Moreover, using the lncRNA SNP database, Wang et al predicted the rs3200401 potential functions, indicating that C to T leads to free energy change which may alter the structural features of MALAT1. The altered MALAT1 may result in weakened interaction between MALAT1 and SRSF2.19 SRSF2 contributed to the tumour phenotype of HPV16-positive cervical cancer cells.32 The SRSF2 depletion leads to the decreased cell proliferation and colony formation, and increased apoptosis.32 SF2/ASF, SRp20 and SRSF2 are upregulated in a cervical cancer progression model, indicating that they may have oncogenic functions.33 Thus, rs3200401 may alter the expression levels of SRSF2 and participate in CC development.
In the current study, we investigated the association of eight SNPs in ANRIL and MALAT1 between healthy control and patients with CC in a Han Chinese population. Our data showed that rs3200401 in MALAT1 and rs4977574 in ANRIL could play key roles in the development of CC. In the current study, one of the limitations should be our study only found rs3200401 in MALAT1 and rs4977574 in ANRIL were associated with the development of CC. However, the function of these two SNPs in the development of CC should be validated in vitro and in vivo. Another limitation was the relatively modest sample size. In the future, large-scale association studies, especially different populations, are required to clarify the role of these SNPs in the susceptibility and development of CC.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was supported by grant from the CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-2-004), National Science Foundation of China (82103190); PUMC Youth Fund (3332019111); Yunnan Province Clinical Research Center for Gynecological and Obstetric Disease (2022YJZX-FC04); Applied Basic Research Projects of Yunnan province (202101AS070205, 202101AU070191); Special Funds for High-level Healthy Talents of Yunnan Province (L-201615, H-2018014 and D-2018037). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Arbyn M, Weiderpass E, Bruni L, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health. 2020;8(2):e191–e203. doi:10.1016/S2214-109X(19)30482-6
2. Gadducci A, Guerrieri ME, Cosio S. Adenocarcinoma of the uterine cervix: pathologic features, treatment options, clinical outcome and prognostic variables. Crit Rev Oncol Hematol. 2019;135:103–114. doi:10.1016/j.critrevonc.2019.01.006
3. Bowden SJ, Bodinier B, Kalliala I, et al. Genetic variation in cervical preinvasive and invasive disease: a genome-wide association study. Lancet Oncol. 2021;22(4):548–557. doi:10.1016/S1470-2045(21)00028-0
4. Chen D, Juko-Pecirep I, Hammer J, et al Genome-wide association study of susceptibility loci for cervical cancer. J Natl Cancer Inst. 2013;105(9):624–633. doi:10.1093/jnci/djt051
5. Leo PJ, Madeleine MM, Wang S, et al. Defining the genetic susceptibility to cervical neoplasia-A genome-wide association study. PLoS Genet. 2017;13(8):e1006866. doi:10.1371/journal.pgen.1006866
6. Shi Y, Li L, Hu Z, et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat Genet. 2013;45(8):918–922. doi:10.1038/ng.2687
7. Takeuchi F, Kukimoto I, Li Z, et al. Genome-wide association study of cervical cancer suggests a role for ARRDC3 gene in human papillomavirus infection. Hum Mol Genet. 2019;28(2):341–348. doi:10.1093/hmg/ddy390
8. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.Can-16-2634
9. Isin M, Dalay N. LncRNAs and neoplasia. Clin Chim Acta. 2015;444:280–288. doi:10.1016/j.cca.2015.02.046
10. Kong Y, Hsieh CH, Alonso LC. ANRIL: a lncRNA at the CDKN2A/B locus with roles in cancer and metabolic disease. Front Endocrinol (Lausanne). 2018;9:405. doi:10.3389/fendo.2018.00405
11. Lu Y, Zhou X, Xu L, Rong C, Shen C, Bian W. Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. Onco Targets Ther. 2016;9:3077–3084. doi:10.2147/ott.S102658
12. Meseure D, Vacher S, Alsibai KD, et al. Expression of ANRIL-polycomb complexes-CDKN2A/B/ARF genes in breast tumors: identification of a two-gene (EZH2/CBX7) signature with independent prognostic value. Mol Cancer Res. 2016;14(7):623–633. doi:10.1158/1541-7786.Mcr-15-0418
13. Qiu JJ, Lin YY, Ding JX, Feng WW, Jin HY, Hua KQ. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol. 2015;46(6):2497–2505. doi:10.3892/ijo.2015.2943
14. Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi:10.3389/fgene.2012.00219
15. Zhang JJ, Wang DD, Du CX, Wang Y. Long noncoding RNA anril promotes cervical cancer development by acting as a sponge of miR-186. Oncol Res. 2018;26(3):345–352. doi:10.3727/096504017X14953948675449
16. Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14(12):1705–1714. doi:10.1080/15476286.2017.1358347
17. Sun Y, Ma L. New insights into long non-coding RNA MALAT1 in cancer and metastasis. Cancers. 2019;11(2):216. doi:10.3390/cancers11020216
18. Miyagawa R, Tano K, Mizuno R, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18(4):738–751. doi:10.1261/rna.028639.111
19. Wang JZ, Xiang JJ, Wu LG, et al. A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: a survival cohort analysis. BMC Cancer. 2017;17(1):167. doi:10.1186/s12885-017-3151-6
20. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–81. doi:10.1093/nar/gkv1340
21. Taheri M, Pouresmaeili F, Omrani MD, et al. Association of ANRIL gene polymorphisms with prostate cancer and benign prostatic hyperplasia in an Iranian population. Biomark Med. 2017;11(5):413–422. doi:10.2217/bmm-2016-0378
22. Qu Y, Shao N, Yang W, Wang J, Cheng Y. Association of polymorphisms in MALAT1 with the risk of esophageal squamous cell carcinoma in a Chinese population. Onco Targets Ther. 2019;12:2495–2503. doi:10.2147/OTT.S191155
23. Yang J, Wang Y, Zhang S, et al. The association of TNF-alpha promoter polymorphisms with genetic susceptibility to cervical cancer in a Chinese Han population. Int J Gen Med. 2022;15:417–427. doi:10.2147/IJGM.S350263
24. Li Z, Zhang Z, He Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 2009;19(4):519–523. doi:10.1038/cr.2009.33
25. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98. doi:10.1038/sj.cr.7290272
26. Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi:10.1093/bioinformatics/btl268
27. Khorshidi HR, Taheri M, Noroozi R, Sarrafzadeh S, Sayad A, Ghafouri-Fard S. ANRIL genetic variants in Iranian breast cancer patients. Cell J. 2017;19(Suppl1):72–78. doi:10.22074/cellj.2017.4496
28. Huang X, Zhang W, Shao Z. Association between long non-coding RNA polymorphisms and cancer risk: a meta-analysis. Biosci Rep. 2018;38(4):BSR20180365. doi:10.1042/BSR20180365
29. Simpson S, Woodworth CD, DiPaolo JA. Altered expression of Erg and Ets-2 transcription factors is associated with genetic changes at 21q22.2-22.3 in immortal and cervical carcinoma cell lines. Oncogene. 1997;14(18):2149–2157. doi:10.1038/sj.onc.1201058
30. Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007;8:39. doi:10.1186/1471-2164-8-39
31. Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi:10.1016/j.molcel.2010.08.011
32. McFarlane M, MacDonald AI, Stevenson A, Graham SV. Human Papillomavirus 16 oncoprotein expression is controlled by the cellular splicing factor SRSF2 (SC35). J Virol. 2015;89(10):5276–5287. doi:10.1128/JVI.03434-14
33. Mole S, McFarlane M, Chuen-Im T, Milligan SG, Millan D, Graham SV. RNA splicing factors regulated by HPV16 during cervical tumour progression. J Pathol. 2009;219(3):383–391. doi:10.1002/path.2608
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
