Back to Journals » International Journal of General Medicine » Volume 13
Association of GSTP1, GSTT1 and GSTM1 Gene Variants with Coronary Artery Disease in Iranian Population: A Case–Control Study
Authors Pourkeramati A , Zare Mehrjardi E , Dehghan Tezerjani M , Seifati SM
Received 5 March 2020
Accepted for publication 1 May 2020
Published 28 May 2020 Volume 2020:13 Pages 249—259
DOI https://doi.org/10.2147/IJGM.S252552
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Alemeh Pourkeramati,1 Ehsan Zare Mehrjardi,1 Masoud Dehghan Tezerjani,2 Seyed Morteza Seifati1
1Medical Biotechnology Research Center, Ashkezar Branch, Islamic Azad University, Ashkezar, Yazd, Iran; 2Abortion Research Centre, Yazd Reproductive Sciences Institute, Shahid Sadoughi University of Medical Science, Yazd, Iran
Correspondence: Seyed Morteza Seifati
Medical Biotechnology Research Center, Ashkezar Branch, Islamic Azad University, Yazd, Iran
Tel +98913-1546072
Email [email protected]
Background: Coronary artery disease (CAD) is a multifactorial disease that may be caused by the interaction between environmental and genetic risk factors. Glutathione S-transferases (GSTs) are known to participate in detoxification and metabolism of a wide range of xenobiotic compounds and oxidative stress products. Considering the interaction between environmental and genetic factors in CAD, we investigated the genetic polymorphisms of GSTM1, GSTT1, and GSTP1 in the Iranian population.
Patients and Methods: Two hundred and forty-four CAD cases and 281 healthy controls were studied. The genotype of GSTM1, GSTT1, and GSTP1 genes was determined by multiplex polymerase chain reaction (PCR) and PCR-restriction fragment length polymorphism (PCR-RFLP) techniques. Multivariable logistic regression analysis was used to calculate the odds ratios (ORs) and 95% confidence intervals (CI). Multifactor dimensionality reduction (MDR) analysis was also carried out to analyze the gene–gene and gene–environment interaction.
Results: The genotype and allele distribution of the three variations were not significantly different between CAD patients and controls (p > 0.05). The subgroup analysis revealed no significant gene–gene interactions or gene–gene combination effects linked to CAD susceptibility. However, MDR analysis selected the GSTM, GSTT pairwise and three genes combination models associated with the susceptibility to CAD. In addition, its result revealed that smoking in combination with GSTM1 (two-way) and GSTT, GSTP (three-way) genes might increase the risk of CAD. Furthermore, a significant interaction between GSTT1-null polymorphism and dyslipidemia was found in multivariable logistic regression analyses in the gene–environmental interactions on CAD risk.
Conclusion: Our results suggest that the GSTM1, GSTT1 and GSTP1 genetic variations are not directly associated with the susceptibility to CAD in Iranian patients. Due to MDR results, there might be a non-linear association between interactions of two or three genes and smoking with CAD. There is also an association between CAD risk factors and GST variations, which requires supplementary confirmation with larger sample sizes.
Keywords: GSTM1, GSTT1, GSTP1, detoxification system, polymorphisms, coronary artery disease
Introduction
Coronary artery disease (CAD) is the most common type of heart disease worldwide, which can lead to a heart attack or death.1 A high prevalence of CAD in the Iranian adult population has indicated.2 CAD is a multifactorial disease affected by both acquired and inherited factors.3 Modifiable risk factors could be ameliorated through lifestyle changes and medication management. However, other risk factors are non-modifiable, such as genetic predisposition and sex.4 It is hypothesized that the development of CAD or atherosclerosis is affected by the interaction between predisposing genes and environmental factors.5 To improve strategies for prevention, early diagnosis, and therapy, the genetic factors involved in CAD development need to be understood.3
There are two main phases associated with detoxification in the eukaryotic cells. In Phase I, cytochrome P450 superfamily of enzymes (CYP450) have a fundamental role in bio transforming xenobiotics, steroid hormones, and pharmaceuticals to highly reactive compounds by adding active groups, including hydroxyl, carboxyl, or an amino group. These procedures through oxidation, reduction, and/or hydrolysis reactions can create reactive electrophilic species, which cause oxidative damage in cellular pathways.6 Phase II detoxification encompasses a procedure, in which an endogenous hydrophilic substance is conjugated to the reactive site of a product from phase I, turning it to a hydrophilic compound increasing its excretion in bile and/or urine. Several enzymes, such as glucuronyl transferases, sulfotransferases, glutathione transferases, amino acid transferases, N-acetyl transferases and N- and O-methyltransferases are involved in this phase.7
Glutathione S-transferase (GST) is one of phase II enzymes, which catalyzes conjugation reactions between glutathione and a wide range of electrophilic substrates. It also detoxifies a wide range of substrates, such as the compounds found in cigarette and environmental contamination that may increase the risk of CAD.8,9 It is also involved in the reactive oxygen species (ROS) reduction and protects the cell against ROS damage.10 The vascular layer including endothelium, smooth muscle and adventitia produce ROS. These molecules act as a signaling molecule under normal physiological condition; however, increased amount of them can cause cell necrosis, apoptosis and dysfunction through oxidizing macromolecules, such as proteins, lipids, and DNA.11 Oxidization of low-density lipoproteins (LDL) is the primary step in atherosclerosis progression, as the main cause of CAD. The oxidized LDL (OxLDL) is involved in several biological procedures, which can be responsible for atherosclerosis in CAD. It increases the activation and proliferation of monocyte and macrophage in the arterial wall through chemotactic activity.12 OxLDL promotes the production of collagen from smooth muscle cells (SMCs), resulting in the formation of the fibrous cap in atherosclerotic plaque and the growth of the lesion.13,14 It also has a cytotoxic effect and increases apoptosis in vascular cells.15,16 Also, OxLDL increases the aggregation and adhesion of platelet by reducing the synthesis of nitric oxide and increasing the production of prostaglandins and related precursors.17,18 Consequently, any changes in the expression of GSTs, responsible for the reduction of ROS, can increase the susceptibility to CAD.
The genetic variants influence the expression and functional activities of the GST proteins. Among the cytosolic GST enzymes, the variations of mu, theta, and pi classes have intensively investigated in various studies.19,20 The glutathione S-transferase mu 1 (GSTM1), glutathione S-transferase theta 1 (GSTT1), and glutathione S-transferase pi 1 (GSTP1) genes are polymorphic and some allelic variants cause enzyme deficiency.9
GSTP1 gene is located on chromosome 11q13 and is expressed in normal epithelial cells, such as cardiovascular system.21 An A to G transition in codon 105 of GSTP1 enzyme leads to the substitution of isoleucine (Ile) to valine (Val) amino acids (Rs 1625).22,23 This alteration affects the enzyme activity compared with the wild-type. Amino acid 105 lies near the active site of the enzyme, by which influences the GSTP1 catalytic activity.24,25
The GSTT1 and GSTM1 encoding genes are organized in gene clusters on chromosomes 22q11 and 1p13.3, respectively.26–28 The common deletion polymorphism of GSTM1 and GSTT1 genes consequently results in the absence of functional enzyme.29,30 Deleting variants or null variants in GSTT1 and GSTM1 genes may arise by homologous recombination of the left- and right-repeated sequences, which results in a 54-kb and 16-kb deletion including the entire two genes.31,32 Regarding the importance of these polymorphic genes, we conducted this study to understand whether the polymorphisms of these three genes or their combinations have any effect on disease progression or susceptibility. Furthermore, another objective of this study was to determine the possible high-level gene–gene and gene–environment interaction between GSTTI, GSTM1, and GSTP1 genes and environmental factors using multifactor dimensionality reduction (MDR) to increase the risk of CAD.
Patients and Methods
Study Population
Two hundred and forty-four unrelated patients with CAD and 281 age- and sex-matched unrelated healthy control subjects were enrolled in this case–control study. The study protocol was approved by the Ethics Committee of Islamic Azad University, Yazd Branch (Ethics code: IR.IAU.REC1396.23). All subjects were given information about the study before their enrollment, and the written informed consent was obtained from patients and healthy individuals. This study was conducted in accordance with the Declaration of Helsinki. Cases were selected randomly from the patients who were referred to the Rajaei Cardiovascular, Medical Research Center, Tehran, Iran from 2011 to 2013. CAD was detected by coronary angiography for patients and defined as stenosis more than 70% in at least one of the major coronary arteries. Patients with concomitant inflammatory or malignant disease were excluded. Control subjects were collected randomly from healthy people who volunteered for the study and examined by a cardiologist. Subjects with concerning signs or symptoms were subjected to coronary angiography to ensure about their health conditions. Subjects with no history of cardiac pain and other CAD risk factors were selected as controls.
Definition of Cardiovascular Risk Factors
The well-known independent risk factors for CAD are hypertension, diabetes, smoking, and dyslipidemia. Hypertension was diagnosed in patients based on a systolic blood pressure (SBP) ≥140 mmHg and/or a diastolic blood pressure (DBP) ≥90 mmHg or the need for blood pressure-lowering medicine.33 Having at least two measurements of fasting blood glucose ≥126 mg/dl was defined as diabetes.34 Smoking was defined as smoking constantly or over repeated periods at least six months. Dyslipidemia was defined as having high levels of blood lipids (eg triglycerides >200 mg/dl, total cholesterol levels ≥240 mg/dl, HDL cholesterol ≤40 mg/dl, LDL cholesterol ≥130 mg/dl) or use of lipid-lowering drugs.35 The Body Mass Index (BMI) was calculated as weight/height2 in all subjects.34
Genotyping
Five mL of peripheral blood samples were collected in Conical Centrifuge Tubes containing EDTA and stored at 4°C. Genomic DNA was isolated using the GenExTM blood genomic DNA purification kit (GeneAll, Korea) according to the experimental protocol. DNA quality and quantity were assessed by the NanoDrop™ 2000 Spectrophotometer.
GSTP1 Genotyping
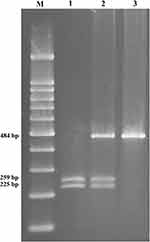
The GSTP1 (Ile105Val) genetic variant was identified using the polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) analysis. A 493-bp fragment included exon 5 substitution was amplified with a set of primers: forward primer: 5ʹ-TCTCATCCTTCCACGCACATC-3ʹ and reverse primer: 5ʹ-TGCTGGAGGTCTCTGTCCTTG-3ʹ. The PCR conditions were 95°C for 5 minutes, 35 cycles of 94°C for 45 seconds, 63°C for 40 seconds and 72°C for 40 seconds, followed by 72°C for 10 minutes. The PCR products were digested with Alw26I (Thermo Scientific™, Norway) at 37°C for 16 h in a total volume of 10 μL. The digestion products were analyzed on a 2% agarose gel prepared in 0.5X TBE buffer. The GSTP1 genotypes were determined as follows: The AA genotype (Ile/Ile) yields two fragments of 484 and 9 bp; the 9 bp is an invariant polymorphism, which used as an internal standard for digestion process; AG genotype (Ile/Val) gives four fragments with 484, 225, 259, and 9 bp and GG mutant genotype (Val/Val) produces three fragments with 225 and 259 bp and 9 bp (Figure 1). After genotyping all samples with PCR-RFLP, some of them were retested by sequencing the PCR product by Macrogen, Korea; no discrepancies were found.
GSTM1 and GSTT1 Genotyping
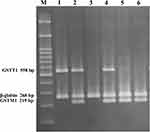
Multiplex PCR was used to detect the GSTM1 and GSTT1 genes in a total volume of 25 μL buffered solution. The reaction mixture was heated at 95°C for 5 min, followed by 35 cycles of amplification as follows: a denaturing step at 94°C for 30 s, an annealing step at 63°C for 1 min, and an extension step at 72°C for 1 min. The final extension was at 72°C for 10 min. PCR samples were analyzed on a 2% agarose gel prepared in 0.5X TBE buffer runs at 110 V for 70 min at room temperature. The absence of a 219-bp band for GSTM1 or a 558-bp band for GSTT1, with the presence of a 268-bp β‑globin (as control fragment) band, was recorded as null genotype (Figure 2). This method did not permit the detection of heterozygous carriers of GSTM1 or GSTT1 deletions, but it identified the null genotypes conclusively. The following primers were used: designed GSTT1 primers: forward: GGTCCTCACATCTCCTTAGC; GSTT1 reverse: AGTCTTAGGCAAGCCATTCC; GSTM1 forward: GAACTCCCTGAAAAGCTAAAGC; GSTM1 reverse: GTTGGGCTCAAATATACGGTGG;36 β‑globin forward: CAACTTCATCCACGTTCACC; β‑globin reverse: GAAGAGCCAAGGACAGGTAC.37
Statistical Analysis
Statistical analysis was carried out using SPSS version 20.0 (SPSS Inc., Chicago, IL, USA). Allele and genotype frequencies in cases and controls were compared using a chi-square (χ2) test, and odds ratios (ORs) and their 95% confidence intervals (CIs) were calculated. Age, the ratio of low-density lipoprotein (LDL) to high-density lipoprotein (HDL), triglycerides and BMI were evaluated by Student’s t-test. The risk factors for CAD and the association between CAD and GSTs polymorphisms were evaluated by multiple logistic regression analysis. Tests for Hardy–Weinberg equilibrium was also conducted by the χ2 test. A p-value <0.05 was defined as statistically significant.
MDR Analysis
Gene–gene interaction and gene–environment interaction analysis between GSTTI, GSTM1, and GSTP1 genes and smoking, as an environmental factor, were executed using the MDR software package (version 3.0.2).38 MDR, as a non-parametric approach, and data mining approach can characterize the gene–gene and gene–environmental nonlinear interaction in a small group and overcome size limitation.38,39 In this analysis, the optimal prediction model was found due to maximum testing balance accuracy (TBA) and cross-validation consistency (CVC). During the analysis, 1000-fold permutation testing and χ2 test at 0.05 significance level were used to test the model with the highest TBA and CVC.
Results
Demographic Information
This study was conducted on 244 patients with CAD and 281 healthy controls. The lifestyle and clinical parameters of the patient and control subjects are summarized in Table 1. No significant difference was found between the case and control groups in age (56.56±9.27 vs 55.46±10.95 years, respectively). Subjects with CAD had a significantly higher frequency of hypertension than controls (45.9% vs 28.4%, respectively, p=0.002), as well as diabetes mellitus (53.7% vs 19.2%, respectively, p=0.000), history of cigarette smoking (46.3% vs 18.8%, respectively, p=0.000), and dyslipidemia (76.6% vs 31.6%, respectively, p=0.000). Male subjects showed a higher rate of CAD (cases: 71.3% vs controls: 58.3%).
 |
Table 1 Clinical Parameters of the Patients and Controls |
CAD patients were classified into three groups based on the CAD severity according to the coronary angiographic results. Of these, 44 (18%), 69 (28.3%), and 131 (53.7%) CAD cases had one-, two-, and three-vessel disease, respectively.
Genotype Frequencies and Their Associations with CAD
The distribution of GSTP1 genotypes between the two groups was not significantly different (p=0.921). The frequency of AA, AG, and GG genotypes among the studied group was 52.9%, 40.9%, and 6.2% in patients; and 50.8%, 42.8%, and 6.4% in the control group, respectively (Table 2). The chi-square test revealed that genotype distributions between patients and controls were not significantly different. The allelic distribution of the rs1625 genotype in the cases and controls showed no deviation from the Hardy–Weinberg equilibrium (p> 0.05).
 |
Table 2 GST Genotypes and the Risk of Coronary Artery Disease |
The proportion of the GSTM1-null genotype was not significantly different between the patients (47.5%) and control (50.9%) groups (p=0.566). This variant did not increase the risk of CAD development. The frequency of the GSTT1-null genotype was 26.2% and 19.9% in the patient and control groups, respectively (p=0.165). No significant association was found between GSTT1 mutation and susceptibility to CAD. Additionally, the distribution of GSTM1 and GSTT1 phenotypes and GSTP1 genotypes in three subgroups of CAD severity is summarized in Table 3.
 |
Table 3 Distribution of the GST Genotypes Stratified by the Severity of Coronary Artery Disease |
The Combined Effect of GST Polymorphisms and Risk of CAD
To further assess the existence of an interaction between the studied GST genes, the combination of their polymorphisms was investigated. The frequencies of these combinations are summarized in Tables 4 and 5. Those who carried all three wild-types (low-risk) genotypes [GSTM1 and GSTT1 (present) and GSTP1 (AA) genotypes] were defined as the reference group (Table 4). No statistically significant association was observed with each genotype combination and the risk of CAD (p=0.192).
 |
Table 4 Combined Effects of GSTM1, GSTT1, and GSTP1 Polymorphisms in the Study Subjects |
 |
Table 5 Combination of Double GST Polymorphisms |
Interactions Between GST Genes and CAD Risk Factors
Supplementary subgroup analyses were performed to determine the interactions between GST genotypes and CAD risk factors (such as hypertension, smoking, dyslipidemia, male gender, and diabetes mellitus) and the risk of CAD development (Table 6). Based on the analysis of the interactions between GSTM1, GSTT1 phenotypes or GSTP1 genotypes and CAD risk factors in CAD pathogenesis, a significant interaction between GSTT1 deletion polymorphism and dyslipidemia on CAD development was observed (p=0.002). No association was observed between and within the other study subgroups.
 |
Table 6 Interaction of the GST Genotypes and Coronary Artery Disease (CAD) Risk Factors on CAD Development |
Gene–Gene and Gene–Environment Interactions by MDR Analysis
The current study also investigated the interaction between GSTTI, GSTM1, GSTP1 genes and smoking as an environmental factor using MDR analysis in controls and CAD patients. The best possible interaction between three genes is listed in Table 7. The GSTT1 gene (null and present genotypes) with the cross-validation of 9/10 had the highest testing-balanced accuracy (51.63%) among the three genes. Among the two-way gene interaction models, the best model was the interaction of GSTM and GSTT genes with a testing-balanced of 47.72% and permutation testing p-value of 0.0254. However, the testing-balanced accuracy of this model is lower compared with the three-way gene interaction model. The three-way genes interaction model showed cross-validation of 10/10 and permutation testing p-value of 0.0087. These results showed that the two-way (GSTM, GSTT) and three-way interaction of three genes might have a non-linear association with the susceptibility to CAD. Figure 3 summarizes the dimensional reduction of the three-way gene interaction of GSTTI, GSTM1, and GSTP1 genes (12 genotypes) showing the high- and low-risk combination of genotypes associated with CAD, as well as the distribution of cases and controls for each combination.
 |
Table 7 Summary of Multifactor Dimensionality Reduction Gene–Gene Interaction Result |
A summary of the best models for two-way and three-way gene-environment interactions between the three genes and smoking are listed in Table 8. These three models have a cross-validation consistency of 100% and significant permutation testing p-value (P<0.0001). The best models for two-way and three-way gene-environment interactions were GSTM-smoking and GSTT-GSTP-smoking, respectively.
 |
Table 8 Summary of Multifactor Dimensionality Reduction Gene–Environment Interaction Results |
Discussion
ROS are the initiator of oxidative stress involved in the pathogenesis of atherosclerosis through several important enzyme systems.40 Risk factors for atherosclerosis and CAD increase the production of ROS by endothelial cells. Although certain enzymes are acting against ROS, such as superoxide dismutase and glutathione peroxidase, these molecules can oxidize lipid, DNA, protein, and carbohydrate, leading to degradation of them and increase their toxicity and mutagenicity. Other enzymes, such as aldehyde dehydrogenase, alcohol dehydrogenase, Aldo-Keto Reductase, and GST neutralize the excessive ROS and the byproducts of oxidative stress. Among these enzymes, GST enzymes catalyze the conjugation of glutathione to a wide variety of electrophile compounds and make them more soluble and less active. Studies also revealed that materials generating free radicals and H2O2 can induce GST in mammalian cells. Therefore, the level of GST expression can be a determining factor for evaluating the response of cells to xenobiotic and chemical compounds, and considered as a potential biomarker for ROS-related diseases.41–43
Many studies have evaluated GST polymorphisms in CAD; however, few studies have addressed the role of GSTM1, GSTT1 and GSTP1 gene polymorphisms in this disease simultaneously.9,44 This study aimed at assessing the association between polymorphisms of GST genes and CAD in the Iranian population. The GSTP1 A1578G transition leads to isoleucine substitution to valine, which affects the enzyme activity. The proportion of Val allele in the control group was 28%. No significant differences were observed in the frequencies of the GSTP1 genotypes between the patient and control groups. Our result is comparable to the allele frequencies in Caucasians (33.1%) and also to a population from the west of Iran (31.9%).44,45 Few studies evaluated the association of GSTP1 polymorphism and CAD,9,44 and our findings are consistent with the results obtained in the west of Iranian and Taiwanese populations. The GSTM1-null polymorphism is caused by homozygous deletion. The frequency of the GSTM1 null genotype in our control subjects was 50.9%, similar to frequencies of this genotype in the Caucasian, Asian, and Iranian control populations (53.1, 52.9, and 50.9%, respectively).46,47 The prevalence of GSTM1-null genotype was 52.8% in a similar study in the west of Iran.44 The frequency of the GSTT1-null genotype in our control participants was 19.9%, which was similar to that reported in the Caucasian and Iranian control population.46,47 The Asian control population showed a higher prevalence of 47% of GSTT1-null genotype.46 The frequency of the GSTT1-null genotype in a control population from the west of Iran was 15.7%.44 In our study, no significant differences were found in the frequencies of GSTM1- and GSTT1-null mutations between the case and control groups as observed in the Taiwanese and Brazilian populations.9,48 In contrast, some studies reported the association between GSTM1- and GSTT1-null genotypes and CAD development in the Italian, Saudi Arabian, North Indian, Chinese, and especially Iranian (west of Iran) populations.44,49-52 A study in the North India found a protective role of the GSTT1-null genotype against CAD.53 Moreover, a study in young South African Indians documented that GSTM1-null genotype is associated with a 2.6-fold higher risk of CAD development.54
Although there was no direct association between the genes and susceptibility to CAD based on traditional statistical analysis, high-level MDR analysis revealed that there might be an association between the interactions of these genes and susceptibility to CAD in our study. Determining the gene–gene and gene–environment interactions are fundamental in epidemiological studies to identify the etiology of different diseases and predict the risk factors for disease prevention.55
Earlier studies investigating the interactions between GST genes and cigarette smoking on CAD disease showed a significant association between GST polymorphisms and CAD risk in smokers.5,56 No direct association was found among the smoker subgroup; however, MDR demonstrated that there might be an association between GSTM and smoking (the best two-way model) and GSTT, GSTP, and smoking (the best three-way model) with the risk of developing CAD in our population. No previously published studies have investigated the interaction between GST genotypes and other CAD risk factors in CAD development. Our results showed that the interaction between GSTT1-null genotype and dyslipidemia significantly affected CAD development (p=0.002). The other interaction between GST genotypes and risk factors for CAD was not significantly different between the two studied groups.
Ethnic differences and varied environmental, lifestyle, cultural, socioeconomics and nutritional factors might explain inconsistent results from different studies.57,58 In terms of ethnicity, it might be determined by self-reported data, which increases its complication. In some population, individuals are inclined to conceal their main ethnicity and use other ethnicity due to the political or cultural sensitivity. Marriage between different ethnic groups can also add more variety to the results of different relevant studies.59 On the other hand, the genetic background and expression of compensatory functions can cover the effect of a gene polymorphism or mutation.25
We observed that the well-known CAD risk factors, like male gender, cigarette smoking, hypertension, dyslipidemia, and diabetes were significantly associated with CAD development. Our data is consistent with the results of previous studies.9,44,54,60
Prior studies have demonstrated conflicting results regarding the combination effect of GSTs genotypes on CAD. Here, we investigated the effects of a combination of two and three genotypes on CAD. The results showed that different combinations of genotypes did not affect CAD. The incidence of a combination of GSTM1- and GSTT1- null genotypes in our control individuals was 8%, which is similar to the previous reports of the frequency of the double nulls between Iranian and Caucasian populations, and a population from the west of Iran (11.8, 10.4 and 10.2%).44 However, in an Asian control population, both GSTM1- and GSTT1-null genotypes were higher (24.6%).46
Conclusion
In conclusion, our study revealed no association between the GSTM1, GSTT1, and GSTP1 genetic variations and the susceptibility to CAD in Iranian patients. However, MDR analysis revealed a two-way interaction between GSTM and GSTT and a three-way combination between GSTT and GSTP of the genes associated with the susceptibility to CAD. In addition, our results revealed that smoking in combination with GSTM1 (two-way) and GSTT and GSTP (three-way) might increase the risk of CAD. There was an association between GSTT1 deletion polymorphism and dyslipidemia, as one of the CAD risk factors on CAD development.
Acknowledgments
The authors are thankful to the patients and healthy individuals for their kind participation.
Disclosure
The authors report no conflicts of interest in this work.
Reference
1. Hanson MA, Fareed MT, Argenio SL, Agunwamba AO, Hanson TR. Coronary artery disease. Prim Care. 2013;40(1):1–16. doi:10.1016/j.pop.2012.12.001
2. Ebrahimi M, Kazemi-Bajestani S, Ghayour-Mobarhan M, Ferns G. Coronary artery disease and its risk factors status in Iran: a review. Iran Red Crescent Med J. 2011;13(9):610. doi:10.5812/kowsar.20741804.2286
3. Zhao Q, Wei H, Liu D, et al. PHACTR1 and SLC22A3 gene polymorphisms are associated with reduced coronary artery disease risk in the male Chinese Han population. Oncotarget. 2017;8(1):658.
4. Zhi H, Wang H, Li T, Pin F. Relationship between GPX1 rs1050450 variation and the onset risk of coronary artery disease. Int J Clin Exp Pathol. 2016;9(2):2325–2329.
5. Song Y, Shan Z, Luo C, et al. Glutathione S-transferase T1 (GSTT1) null polymorphism, smoking, and their interaction in coronary heart disease: a comprehensive meta-analysis. Heart Lung Circ. 2017;26(4):362–370. doi:10.1016/j.hlc.2016.07.005
6. Danielson P. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3(6):561–597. doi:10.2174/1389200023337054
7. Xu C, Li CY-T, Kong A-NT. Induction of Phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28(3):249. doi:10.1007/BF02977789
8. Kelly FJ, Fussell JC. Role of oxidative stress in cardiovascular disease outcomes following exposure to ambient air pollution. Free Radic Biol Med. 2017;110:345–367. doi:10.1016/j.freeradbiomed.2017.06.019
9. Yeh H-L, Kuo L-T, Sung F-C, Chiang C-W, Yeh -C-C. GSTM1, GSTT1, GSTP1, and GSTA1 genetic variants are not associated with coronary artery disease in Taiwan. Gene. 2013;523(1):64–69. doi:10.1016/j.gene.2013.02.052
10. Nimse SB, Pal D. Free radicals, natural antioxidants, and their reaction mechanisms. RSC Adv. 2015;5(35):27986–28006. doi:10.1039/C4RA13315C
11. Ray PD, Huang B-W, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24(5):981–990. doi:10.1016/j.cellsig.2012.01.008
12. Quinn MT, Parthasarathy S, Steinberg D. Lysophosphatidylcholine: a chemotactic factor for human monocytes and its potential role in atherogenesis. PNAS. 1988;85(8):2805–2809. doi:10.1073/pnas.85.8.2805
13. Jimi S, Saku K, Uesugi N, Sakata N, Takebayashi S. Oxidized low density lipoprotein stimulates collagen production in cultured arterial smooth muscle cells. Atherosclerosis. 1995;116(1):15–26. doi:10.1016/0021-9150(95)05515-X
14. Rajavashisth TB, Liao JK, Galis ZS, et al. Inflammatory cytokines and oxidized low density lipoproteins increase endothelial cell expression of membrane type 1-matrix metalloproteinase. J Biol Chem. 1999;274(17):11924–11929. doi:10.1074/jbc.274.17.11924
15. Cathcart MK, Morel DW, Chisolm GM
16. Sata M, Walsh K. Oxidized LDL activates fas-mediated endothelial cell apoptosis. J Clin Invest. 1998;102(9):1682–1689. doi:10.1172/JCI3531
17. Li L-X, Chen J-X, Liao D-F, Yu L. Probucol inhibits oxidized-low density lipoprotein-induced adhesion of monocytes to endothelial cells by reducing P-selectin synthesis in vitro. Endothelium. 1998;6(1):1–8. doi:10.3109/10623329809053400
18. Armstrong D. Oxidized LDL, ceroid, and prostaglandin metabolism in human atherosclerosis. Med Hypotheses. 1992;38(3):244–248. doi:10.1016/0306-9877(92)90103-J
19. Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482(1):21–26. doi:10.1016/S0027-5107(01)00206-8
20. Moosavi A, Forat YM, Dehghan TM, et al. Glutathione S-transferase M1-T1 null genotypes and susceptibility to Hodgkin’s lymphoma. Genetika. 2017;49(3):911–920. doi:10.2298/GENSR1703911M
21. Zhao E, Zhao Y, Yang Q. Lack of association between GSTP1 Ile105Val polymorphism and coronary heart disease risk: a meta-analysis. Int J Clin Exp Med. 2015;8(10):18488.
22. Zhao R, Chen G. Role of GSTP1 Ile105Val and XRCC1 Arg194Trp, Arg280His and Arg399Gln gene polymorphisms in the clinical outcome of advanced non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(11):14909.
23. Jeon MJ, Choi YM, Hong MA, et al. No association between the GSTP1 exon 5 polymorphism and susceptibility to advanced stage endometriosis in the Korean population. Am J Reprod Immunol. 2010;63(3):222–226. doi:10.1111/j.1600-0897.2009.00780.x
24. Holley SL, Fryer AA, Haycock JW, Grubb SE, Strange RC, Hoban PR. Differential effects of glutathione S-transferase pi (GSTP1) haplotypes on cell proliferation and apoptosis. Carcinogenesis. 2007;28(11):2268–2273. doi:10.1093/carcin/bgm135
25. Vasieva O. The many faces of glutathione transferase pi. Curr Mol Med. 2011;11(2):129–139. doi:10.2174/156652411794859278
26. Thorn CF, Ji Y, Weinshilboum RM, Altman RB, Klein TE. PharmGKB summary-very important pharmacogene information for GSTT1. Pharmacogenet Genomics. 2012;22(8):646. doi:10.1097/FPC.0b013e3283527c02
27. Martins J, Rodrigues D, Silva K, et al. Molecular analysis of the GSTT1 gene polymorphism in patients with clinical manifestation of atherosclerosis. Genet Mol Res. 2017;16(3). doi:10.4238/gmr16039620.
28. Yang M, Zhao J, Xing L, Shi L. Association between GSTM1 null genotype and coronary artery disease risk: a meta-analysis. Med Sci Monit. 2014;20:1550. doi:10.12659/MSM.890876
29. Josephy PD. Genetic variations in human glutathione transferase enzymes: significance for pharmacology and toxicology. Hum Genomics Proteomics. 2010;2010.
30. Hur SE, Lee JY, Moon H-S, Chung HW. Polymorphisms of the genes encoding the GSTM1, GSTT1 and GSTP1 in Korean women: no association with endometriosis. Mol Hum Reprod. 2005;11(1):15–19. doi:10.1093/molehr/gah127
31. Parl FF. Glutathione S-transferase genotypes and cancer risk. Cancer Lett. 2005;221(2):123–129. doi:10.1016/j.canlet.2004.06.016
32. Malik SS, Kazmi Z, Fatima I, Shabbir R, Perveen S, Masood N. Genetic polymorphism of GSTM1 and GSTT1 and risk of prostatic carcinoma—a meta-analysis of 7281 prostate cancer cases and 9082 healthy controls. Asian Pac J Cancer Prev. 2016;17(5):2629–2635.
33. Franklin SS, Jacobs MJ, Wong ND, Gilbert J, Lapuerta P. Predominance of isolated systolic hypertension among middle-aged and elderly US hypertensives. Hypertension. 2001;37(3):869–874. doi:10.1161/01.HYP.37.3.869
34. Fox CS, Golden SH, Anderson C, et al. Update on prevention of cardiovascular disease in adults with type 2 diabetes mellitus in light of recent evidence. Circulation. 2015;132(8):691–718. doi:10.1161/CIR.0000000000000230
35. Kavey R-EW, Daniels SR, Lauer RM, Atkins DL, Hayman LL, Taubert K. American Heart Association guidelines for primary prevention of atherosclerotic cardiovascular disease beginning in childhood. Circulation. 2003;107(11):1562–1566. doi:10.1161/01.CIR.0000061521.15730.6E
36. Hezova R, Bienertova-Vasku J, Sachlova M, et al. Common polymorphisms in GSTM1, GSTT1, GSTP1, GSTA1 and susceptibility to colorectal cancer in the Central European population. Eur J Med Res. 2012;17(1):17. doi:10.1186/2047-783X-17-17
37. López-Guerrero JA, Pellín A, Noguera R, Carda C, Llombart-Bosch A. Molecular analysis of the 9p21 locus and p53 genes in Ewing family tumors. Lab Invest. 2001;81(6):803. doi:10.1038/labinvest.3780290
38. Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene–gene and gene–environment interactions. J Bioinform. 2003;19(3):376–382. doi:10.1093/bioinformatics/btf869
39. Manuguerra M, Matullo G, Veglia F, et al. Multi-factor dimensionality reduction applied to a large prospective investigation on gene–gene and gene–environment interactions. Carcinogenesis. 2007;28(2):414–422. doi:10.1093/carcin/bgl159
40. Harrison D, Griendling KK, Landmesser U, Hornig B, Drexler H. Role of oxidative stress in atherosclerosis. Am J Cardiol. 2003;91(3):7–11. doi:10.1016/S0002-9149(02)03144-2
41. Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45(1):51–88. doi:10.1146/annurev.pharmtox.45.120403.095857
42. Eaton DL, Bammler TK. Concise review of the glutathione S-transferases and their significance to toxicology. Toxicol Sci. 1999;49(2):156–164. doi:10.1093/toxsci/49.2.156
43. Dasari S, Ganjayi MS, Meriga B. Glutathione S-transferase is a good biomarker in acrylamide induced neurotoxicity and genotoxicity. Interdiscip Toxicol. 2018;11(2):115. doi:10.2478/intox-2018-0007
44. Nomani H, Mozafari H, Ghobadloo SM, et al. The association between GSTT1, M1, and P1 polymorphisms with coronary artery disease in Western Iran. Mol Cell Biochem. 2011;354(1–2):181–187. doi:10.1007/s11010-011-0817-2
45. Lewis SJ, Cherry NM, Niven RM, Barber PV, Povey AC. GSTM1, GSTT1 and GSTP1 polymorphisms and lung cancer risk. Cancer Lett. 2002;180(2):165–171. doi:10.1016/S0304-3835(02)00028-9
46. Garte S, Gaspari L, Alexandrie A-K, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10(12):1239–1248.
47. Moasser E, Kazemi-Nezhad SR, Saadat M, Azarpira N. Study of the association between glutathione S-transferase (GSTM1, GSTT1, GSTP1) polymorphisms with type II diabetes mellitus in southern of Iran. Mol Biol Rep. 2012;39(12):10187–10192. doi:10.1007/s11033-012-1893-4
48. Bazo AP, Salvadori D, Salvadori RA, et al. DNA repair gene polymorphism is associated with the genetic basis of atherosclerotic coronary artery disease. Cardiovasc Pathol. 2011;20(1):e9–e15. doi:10.1016/j.carpath.2009.12.004
49. Masetti S, Botto N, Manfredi S, et al. Interactive effect of the glutathione S-transferase genes and cigarette smoking on occurrence and severity of coronary artery risk. J Mol Med. 2003;81(8):488–494. doi:10.1007/s00109-003-0448-5
50. Abu-Amero KK, Al-Boudari OM, Mohamed GH, Dzimiri N. T null and M null genotypes of the glutathione S-transferase gene are risk factor for CAD independent of smoking. BMC Med Genet. 2006;7(1):38. doi:10.1186/1471-2350-7-38
51. Bhat MA, Gandhi G. Association of GSTT1 and GSTM1 gene polymorphisms with coronary artery disease in North Indian Punjabi population: a case-control study. Postgrad Med J. 2016;92(1094):
52. Wang L-S, Tang -J-J, Tang N-P, et al. Association of GSTM1 and GSTT1 gene polymorphisms with coronary artery disease in relation to tobacco smoking. Clin Chem Lab Med. 2008;46(12):1720–1725. doi:10.1515/CCLM.2008.353
53. Girisha K, Gilmour A, Mastana S, et al. T1 and M1 polymorphism in glutathione S-transferase gene and coronary artery disease in North Indian population. Indian J Med Sci. 2004;58(12):520.
54. Phulukdaree A, Khan S, Moodley D, Chuturgoon AA. GST polymorphisms and early-onset coronary artery disease in young South African Indians. S Afr Med J. 2012;102(7):627–630. doi:10.7196/SAMJ.5520
55. Ottman R. Gene–environment interaction: definitions and study designs. Prev Med. 1996;25(6):764. doi:10.1006/pmed.1996.0117
56. Mir R, Bhat MA, Javaid J, et al. Glutathione S-transferase M1 and T1 (rs4025935 and rs71748309) null genotypes are associated with increased susceptibility to coronary artery disease in Indian populations. Acta Cardiol. 2016;71(6):678–684. doi:10.1080/AC.71.6.3178186
57. Amer MA, Ghattas MH, Abo-ElMatty DM, Abou-El-Ela SH. Evaluation of glutathione S-transferase P1 genetic variants affecting type-2 diabetes susceptibility and glycemic control. Arch Med Sci AMS. 2012;8(4):631. doi:10.5114/aoms.2012.30286
58. Lanza G. Ethnic Variations in Acute Coronary Syndromes. BMJ Publishing Group Ltd; 2004.
59. Abbasi SH, Sundin Ö, Jalali A, Soares J, Macassa G. Ethnic differences in the risk factors and severity of coronary artery disease: a patient-based study in Iran. J Racial Ethn Health Disparities. 2018;5(3):623–631. doi:10.1007/s40615-017-0408-3
60. Tamer L, Ercan B, Camsarı A, et al. Glutathione S-transferase gene polymorphism as a susceptibility factor in smoking-related coronary artery disease. Basic Res Cardiol. 2004;99(3):223–229. doi:10.1007/s00395-004-0465-8
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.