Back to Journals » OncoTargets and Therapy » Volume 12
Association Of GSTM1, GSTT1 And GSTP1 Polymorphisms With Breast Cancer Among Jordanian Women
Authors AL-Eitan LN , Rababa'h DM, Alghamdi MA , Khasawneh RH
Received 1 March 2019
Accepted for publication 10 September 2019
Published 20 September 2019 Volume 2019:12 Pages 7757—7765
DOI https://doi.org/10.2147/OTT.S207255
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Faris Farassati
Laith N AL-Eitan,1,2 Doaa M Rababa’h,1 Mansour A Alghamdi,3 Rame H Khasawneh4
1Department of Applied Biological Sciences, Jordan University of Science and Technology, Irbid 22110, Jordan; 2Department of Biotechnology and Genetic Engineering, Jordan University of Science and Technology, Irbid 22110, Jordan; 3Department of Anatomy, College of Medicine, King Khalid University, Abha, Saudi Arabia; 4Department of Hematopathology, King Hussein Medical Center (KHMC), Jordanian Royal Medical Services (RMS), Amman 11118, Jordan
Correspondence: Laith N AL-Eitan
Jordan University of Science and Technology, P.O. Box 3030, Irbid 22110, Jordan
Tel +962-2-7201000 Ext 23464
Fax +962-2-7201071
Email [email protected]
Purpose: Genetic predisposition to disease has become one of the most investigated risk factors in recent years, and breast cancer (BC) is no exception. In this study, we investigated specific genetic variants of three candidate genes belonging to the glutathione-S-transferase superfamily that have been implicated in increased risk of cancers.
Materials and methods: This case-control study comprised 241 Jordanian women who were diagnosed with BC in addition to 219 matched controls. Gel electrophoresis of PCR products was used to visualize and genotype both the GSTM1 and GSTT1 genes, while PCR-RFLP was employed to genotype the rs1695 of the GSTP1 gene.
Results: Our findings did not reveal any correlation between the investigated polymorphisms of GST genes and BC risk among Jordanian women. Otherwise, the combination of GSTM1 entire gene deletion and (GG) genotype of GSTP1 polymorphism (rs1695) was significantly associated with BC with p-value <0.05 (i.e. p-value was not significant after correcting for multiple comparison).
Conclusion: We suggest that the interaction between GSTM1 polymorphism and rs1695 of GSTP1 may influence BC development and progression among Jordanian women. More epidemiological studies are needed to provide a baseline for the underlying role of GSTs polymorphisms in tumorigenesis.
Keywords: breast, cancer, genetic variation, GST genes, polymorphisms
Introduction
Breast cancer (BC) occurs when the multiplication of specific cells in the breast becomes abnormal and uncontrollable, leading to tumor formation.1 It accounts for 16% of invasive cancers in women worldwide and is responsible for 18.2% of all cancer deaths.2 Despite the high mortality rate, BC incidence and survival rates vary between developed and developing countries, as evidenced by the fact that North American survival rates exceed 80% compared to those that fall below 40% in low-income countries.3 As well, BC is the most common female cancer in Jordan, accounting for 37.3% of all cancers in Jordanian women.4
A range of different factors contribute to increasing the risk of developing BC including an individual’s lifestyle, environment, and genetic makeup.5 Nonetheless, the bulk of BC research has been directed towards the role of inherited factors, mainly genetic polymorphisms in critical genes, in the development of this particular form of cancer.6
Glutathione S-transferases (GSTs) are a family of essential Phase II detoxification enzymes that protect against oxidative stress by catalyzing the conjugation of glutathione and xenobiotic compounds.7 The soluble dimeric is one of the GSTs superfamilies that found mainly in the cytosol and comprise eight classes d Alpha, Kappa, Mu, Pi, Sigma, Theta, Zeta and Omega. Individuals defect the expression of GST-M1, GST-T1 and GST-P1 enzymes are more susceptible to different types of cancers such as breast, colorectal and lung cancers.8 Three common polymorphic genes of this family (GSTM1, GSTT1, and rs 1695 of GSTP1) have been widely investigated in the context of BC research.9 It has previously been reported that whole gene deletion of both the GSTM1 and GSTT1 genes in addition to the rs1695 of GSTP1 gene could be responsible for BC progression.10 rs1695 is a single nucleotide polymorphism (SNP) at codon 105 in the GSTP1 gene that is responsible for the substitution of isoleucine amino acid with valine. The degree of involvement of the aforementioned GST genetic variants with BC varies among different ethnic groups.11 In this regard, a study by Helzlsouer et al revealed that the deletion of both GSTM1 and GSTT1 genes as well as the (AG) genotype of GSTP1 (rs1965) might be associated with BC risk among White Americans.12 In contrast, another study conducted on the Thai population indicated no significant influence of the GSTM1 and GSTT1 deletion genotypes and the rs1695 of GSTP1 on BC progression.13
This study focus on three polymorphic well-identified genes GSTM1 (deletion), GSTT1 (deletion) and GSTP1 (rs1695) in which polymorphisms were extensively investigated with respect to cancer in molecular epidemiologic research. The variability in GST function is attributed to the polymorphisms in these GST. Therefore, characterizing the frequencies of these polymorphisms within Jordanian population from Arab descent and compare it to other population may help to understand the exact role of GST in cancer development and progression.
Materials And Methods
Study Population
This case-control study included 241 patients diagnosed with BC in addition to 219 unrelated healthy females with no family history of BC disease and was approved by the Human Ethics Committee at Jordan University of Science and Technology. Samples from Jordanian BC patients were collected from the Jordanian Royal Medical Services (JRMS) hospital. In addition, 219 samples from randomly selected healthy Jordanian women were recruited from the blood bank at JRMS. Both the patients and the controls were age- and gender-matched and came from the same ethnic background (Arab).
GSTM1, GSTT1, And GSTP1 (rs1695) Genotyping
For each sample, the Wizard® Genomic DNA Purification Kit (Promega Corp., Madison, WI, USA) was used to extract genomic DNA from 5 mL of blood according to the manufacturer’s instruction. DNA quantity (ng/µL) and purity (A260/280) were verified using the NanoDrop™ spectrophotometer. For both the GSTM1 and GSTT1 genes, genotypes were assessed by detecting the presence or absence of each gene.
Traditional PCR was carried out to genotype each gene using specific sets of primers (Table 1).14 PCR protocol was conducted in the following manner: 12.5 µL of ready-made master mix plus 10 µL of deionized water were added to a 25 µL reaction tube, followed by 2 µL of each forward and reverse primer in addition to 50 ng of the purified DNA.
 |
Table 1 Primer Sequences And Predicted Product Sizes Of PCR Amplified Products Of The GST Genetic Variants |
The amplified conditions for GSTM1 presence, GSTT1 presence, GSTT1 deletion and the (rs1695) of GSTP1 (A/G) were carried out as previously described.15 On the other hand, GSTM1 deletion was detected by a PCR program involving an initial denaturation of 3 mins, 35 cycles of denaturation (30 s of at 94°C), annealing (1 min at 64.5°C), and extension (1:30 mins at 68°C), and a final extension for 10 mins at 72°C. PCR products were separated using 1.5% agarose gel and then visualized under UV light. SNP detection within the GSTP1 gene was done by using PCR followed by restriction fragment length polymorphism (RFLP).16
Statistical Analysis
The Hardy–Weinberg equilibrium equation was used to calculate the genotypic and allelic frequencies, while Pearson’s chi-squared and ANOVA tests were used to perform the genetic association analysis. In terms of P values, the level of significance was taken as P<0.05. The Statistical Package for the Social Sciences (SPSS) version 21.0 was used to perform all analyses.
Correction For Multiple Testing
Method of Li and Ji (2005) was used to estimate the effective number of genetic variants (Nem),17 which employs a modification of an earlier approach by Nyholt (2004).18 Modified Bonferroni procedure was applied to determine a target alpha level (0.05/Nem) that would maintain an overall significance level of 0.05 or less.
Results
Sample Characteristics
This study involved 241 breast cancer female patients selected from Jordanian population in addition to 219 healthy subjects. Both the cases and controls were randomly chosen and adjusted to be matched with regard to age, sex, and ethnic origin and were all 100% native Arab ancestry (genetically homogenous). The study cohorts were analyzed and summarized in previously published study.4 However, the demographical clinical and pathological data were available for 230 patients. Briefly, patients’ ages ranged from 24 to 95 with the average (±SD) of 53.9 ± 12.777 years, while the average age of controls was 50.4 ± 12.607 years and ranged from 24 to 90 years.
Clinico-Pathologic Features Of Breast Cancer (BC) Patients
Table 2 shows several features of BC including clinical and pathological. The majority (67.89%) of participants were older than 45 years old at first diagnosis with BC. In addition, most of the patients had gotten pregnant and breastfed from an early age (less than 20 years old) (83%), while 72.49% of the cases experienced menarche at an age older than 13 years old. Furthermore, pathological parameters including histopathological characteristics, progesterone (PR) and estrogen receptors (ER) status, lymph node involvement, axillary lymph nodes metastatic, and tumor size were considered in this study. We found that 76.7% of the cases expressed the ER receptor while 48.9% of them were PR positive. Moreover, 90% of patients were diagnosed with low tumor grade compared to 10% who had high grade of tumor. The tumor size of patient was extracted from their medical records, we estimated that 74% of BC patients had tumor size of more than 2 cm. Our results revealed that 51.3% of the patients had axillary lymph nodes free of tumor, whereas 48.67% of the BC patients were diagnosed with metastatic carcinoma in the axillary lymph nodes.
 |
Table 2 Clinical And Pathological Features Of BC Patients (n = 241) |
DNA Genotyping And Gel Electrophoresis
The gel electrophoresis technique was used to detect the genotype for the GSTM1, GSTT1, and GSTP1 genetic variants. GSTM1 genotyping was based on the detection of a 625 bp band of one or both gene copies while the 4748 bp band indicated the absence of one or both gene copies. Figure 1 exhibits the three genotypes for the GSTM1 gene. As for GSTM1, the GSTT1 gene was genotyped in the same manner. Figure 2 demonstrates the three genotypes for the GSTT1 gene.
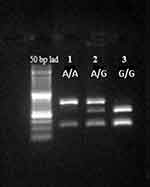
The amplicon size of the GSTP1 gene before digestion was 436 bp. A PCR-RFLP assay was used to detect the SNP within the GSTP1 gene via the BSMA1 restriction digestion enzyme. Incubating the PCR product with this enzyme gave three different genotypes displayed by gel electrophoresis: (A/A), (A/G), and (G/G). Figure 3 illustrates these genotypes and the corresponding band sizes for each one.
Frequency Distribution And Genetic Association Of GSTM1, GSTT1 And GSTP1 Polymorphism With Breast Cancer (BC)
Genotypic and allelic frequencies were statistically analyzed. Table 3 shows the genotypic and allelic distributions of the GSTM1, GSTT1, and GSTP1 (rs1695) polymorphisms among Jordanian BC patients and controls. Our results indicated that the frequency distributions of the double deletion of GSTM1 and GSTT1 genes among Jordanians of Arab descent were 53.4% and 26.9%, respectively, while it was 8.2% for the rs1695 mutant genotype (G/G) within the GSTP1 gene. Allelic distribution for the GSTM1, GSTT1, and rs1695 of GSTP1 polymorphisms also revealed no statistically significant difference between patients and controls.
 |
Table 3 Genotypic And Allelic Distributions Of The GSTM1, GSTT1, And GSTP1 (rs1695) Polymorphisms In Jordanian BC Patients (n = 241) And Controls (n = 219) |
Remarkably, the heterozygous genotype frequency (presence/deletion) for the GSTT1 gene was more frequent than the each of homozygous presence and homozygous deletion genotypes among cases and controls. Furthermore, our finding revealed that the distribution of GSTM1 double deletion genotype was slightly higher among BC patients (57.7%) than it among controls (53.4%). However, we did not find any correlation between each of GSTM, GSTT1 and rs1695 of GSTP1 and BC risk (P = 0.226, 0.590 and 0.659), respectively (Table 3).
Moreover, we investigated the influence of the combination genotypes between the GSTM1, GSTT1, and rs1695 of GSTP1 on BC risk. As shown in Table 4, we estimated a correlation between a (combined GSTM1 homozygous presence with GSTP1 variant genotypes) and BC risk (P = 0.32). We also detected a statistically significant association of combined GSTM1 heterozygous (presence/deletion) and GSTP1 (rs1695) genotypes with BC risk (P = 0.021). However, we propose that the combination between GSTT1 genotypes and GSTP1 (rs1695) was not involved in BC development or progression (Table 4).
 |
Table 4 Distributions of the Combination Genotypes of GSTM1, GSTT1 and Rs1695 of GSTP1 Polymorphisms among Jordanian BC Patients (n = 241) and Controls (n = 219) |
Furthermore, we inspected the genetic association of the investigated GSTs with BC using different genetic models. Table 5 illustrates the different categories of the test and also indicates the chi-squared values. In this study, there was no significant difference between patients and controls for each tested category.
 |
Table 5 Genetic Association Analysis Of GSTM1, GSTT1, And GSTP1 (1695) In BC Patients And Controls Using Different Genetic Models |
Discussion
BC is a disease that influenced by both genetic and environmental factors. BC susceptibility genes are responsible for the development of 20% to 25% of all BC cases.19 GSTs, which take part in the cell’s detoxification process, comprise three common genes (GSTM1, GSTT1 and GSTP1) that are suggested to be involved in BC progression.20 BC is a spectrum of many subtypes with distinct biological characteristic. To understand BC treatment personalization, more accomplished analysis and evaluation of the molecular characteristics of the disease in the individual patient are required. However, the interaction between the genetic variants and BC should be more specific because of the stratified etiology of BC.
Studies have shown that specific genetic variants of these three genes (double deletion for both the GSTM1 and GSTT1 genes neither the rs1695 within the GSTP1 gene) are involved in BC risk.11,12,21 In contrast, other studies have reported that the aforementioned variants are not associated with an increased risk of BC.22–24 Table 6 reviews the most important studies regarding the relationship between BC and the candidate gene variants in different ethnic groups.
 |
Table 6 Genetic Association Studies Of Breast Cancer (BC) Among Different Populations |
The present study’s cohort involved 241 female patients and 219 unrelated healthy female controls. It was revealed that 53.4% of the healthy Jordanian population does not have any copy of the GSTM1 gene, a figure which is close to another finding by previous study within the same population.15 Meanwhile, 57.7% of all patients did not express both GSTM1 copies, which is comparable to findings by Yang et al (2004) in Shanghai (55.8%) but different from findings in a Thai population (35.0%).13 On the other hand, the frequency of the GSTT1 double deletion polymorphism among BC patients was 24.4%, which is incompatible to what was found in the Brazilian (58.8%),1 Thai (41.9%),13 and Californian (82%) populations.10 In this study, we proposed that both GSTM1 and GSTT1 genes were not significantly related to BC risk among Jordanian women.
On the other hand, the frequency of the GSTP1 (AG) genotype was also estimated as a part of the present study. Interestingly, we found that the mutant genotype (GG) among controls (8.2%) is slightly higher than it is in patients (7.9%). Nevertheless, there was no statistical association between the GSTP1 (GG) genotype and increased BC susceptibility, which is in concordance with the frequencies in the Thai population but in contrast to those in the Turkish,25 Chinese,26 and Washingtonian populations.12 In this work, we deduce that the rs1695 of GSTP1 gene was not involved in BC risk among Jordanian females.
Additionally, the association of the combined GSTM1 and GSTT1 polymorphisms with rs1695 of GSTP1 genotypes with BC risk was conducted. In this regard, there are only few studies that inspect the interaction between GST genetic variants within different genes. While the GSTM1 polymorphism alone was not related to BC, our results demonstrated that the combined GSTM1 and GSTP1 (rs1695) genotypes might influence BC risk among Jordanian females, which is contrary to reports for the Chinese population.26 A possible explanation for the divergence between findings among different studies inconsistent results could be that other members of the GST family or other enzymes involved in similar chemical detoxification compensate for the deletion of a functional GSTM1 enzyme. In addition, this gene has not face a strong environmental selection pressure during evaluation.27,28 However, the genetic interaction between the GSTT1 and GSTP1 polymorphisms was not related to BC risk among Jordanian women. Comprehensive epidemiological studies regarding the genetic variations of GSTs among populations and ethnicity are key for implementing the individualization of treatment depending on individual genetic background.
Much significance can be derived from these results, as they help shift the focus of cancer therapy from blanket treatments to more efficient protocols tailored to an individual’s genetic makeup. It can be concluded from the results of the present study that there is no relationship between the genetic variants among the studied GST genes (GSTM1, GSTT1, and rs1695 of GSTP1) and BC risk. However, we proposed that the combination between GSTM1 and GSTP1 polymorphism may be implicated in BC development and progression among Jordanian females of Arab descent. Bearing in mind the accelerated rates of BC incidence around the world, more studies should focus on the nature of this disease in developing countries especially so as to adequately address the impact of the genetic etiology of BC.
Ethical Statement
Ethical approval was obtained from the Intuitional Review Board (IRB) at Jordan University of Science and Technology. Written informed consent was obtained from all participants in this study. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Acknowledgments
The authors thank the Royal Medical Services for approving the study in the first instance.
This study was funded by the Deanship of Research (126/2017), Jordan University of Science and Technology.
Disclosure
The authors report no conflicts of interest in this work.
References
1. U.S. National Library of Medicine. Genetics home reference. Available from: https://ghr.nlm.nih.gov/condition/breast-cancer/; 2017
2. Brophy JT, Keith MM, Watterson A, et al. Breast cancer risk in relation to occupations with exposure to carcinogens and endocrine disruptors: a Canadian case–control study. Environ Health. 2012;11:87. doi:10.1186/1476-069X-11-87
3. World health organization. Cancer. Breast cancer: prevention and control. 2014. Available from: http://www.who.int/cancer/detection/breastcancer/en/.
4. AL-Eitan L, Jamous R, Khasawneh R. Candidate gene analysis of breast cancer in the Jordanian population of Arab descent: a case-control study. Cancer Invest. 2017. doi:10.1080/07357907.2017.1289217
5. Laamiri FZ, Hasswane N, Kerbach A, et al. Risk factors associated with a breast cancer in a population of Moroccan women whose age is less than 40 years: a case control study. Pan Afr Med J. 2016;24:19. doi:10.11604/pamj.2016.24.19.8784
6. Steel M, Thompson A, Clayton J. Genetic aspects of breast cancer. Br Med Bull. 1991;47:504–518. doi:10.1093/oxfordjournals.bmb.a072488
7. Jancova P, Anzenbacher P, Anzenbacherova E. Phase II drug metabolizing enzymes. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2010;154(2):103–116. doi:10.5507/bp.2010.017
8. Luo W, Kinsey M, Schiffman J, Lessnick S. Glutathione S-transferases in pediatric cancer. Front Genet. 2011;1:39.
9. Nebert DW, Vasiliou V. Analysis of the glutathione S-transferase (GST) gene family. Hum Genomics. 2004;1(6):460–464. doi:10.1186/1479-7364-1-6-460
10. Woldegiorgis S, Ahmed RC, Zhen Y, Erdmann CA, Russell ML, Goth-Goldstein R. Genetic Polymorphism in Three Glutathione S-transferase Genes and Breast Cancer Risk. California Breast Cancer Research Program of the University of California; 2002:1–14.
11. Possuelo L, Peraça C, Eisenhardt M, et al. Polymorphisms of GSTM1 and GSTT1 genes in breast cancer susceptibility: a case-control study. Rev Bras Ginecol Obstet. 2013;35(12):569–574.
12. Helzlsouer K, Selmin O, Huang H, et al. Association between glutathione S-transferase M1, P1, and T1 genetic polymorphisms and development of breast cancer. J Natl Cancer Inst. 1998;90(7):512–518. doi:10.1093/jnci/90.7.512
13. Pongtheerat T, Treetrisool M, Purisa W. Glutathione S-transferase polymorphisms in breast cancers of Thai patients. Asia-Pac J Cancer Prev. 2009;10:127–132.
14. Buchard A, Sanchez J, Dalhoff K, Morling N. Multiplex PCR detection of GSTM1, GSTT1, and GSTP1 gene variants. J Mol Diagn. 2007;9(5):612–617. doi:10.2353/jmoldx.2007.060107
15. AL-Eitan L, Rababa’h D, Alkhatib R, Khasawneh R, ALjarrah O. GSTM1and GSTP1 genetic polymorphisms and their associations with acute lymphoblastic leukemia susceptibility in a Jordanian population. J Pediatr Hematol Oncol. 2016;38(7):223–229. doi:10.1097/MPH.0000000000000609
16. Sailaja K, Surekha D, Nageswara D, Rao D, Vishnupriya S. Association of the GSTP1 gene (Ile105Val) polymorphism with chronic myeloid leukemia. Asia-Pac J Cancer Prev. 2010;11:461–464.
17. Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi:10.1038/sj.hdy.6800717
18. Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi:10.1086/382659
19. Alshareeda AT, Negm OH, Albarakati N, et al. Clinicopathological significance of KU70/KU80, a key DNA damage repair protein in breast cancer. Breast Cancer Res Treat. 2013;139:301–310. doi:10.1007/s10549-013-2542-x
20. Zheng T, Holford TH, Zahm SH, et al. Glutathione S-transferase M1 and T1 genetic polymorphisms, alcohol consumption and breast cancer risk. Br J Cancer. 2003;88:58–62. doi:10.1038/sj.bjc.6600708
21. Park S, Kang D, Park B, et al. A case-control study of the association between glutathione S-transferase (GST) M1 and T1 genetic polymorphism and breast cancer in korean women: preliminary report. J Korean Cancer Assoc. 1999;31(4):653–662.
22. Linhares J, Da Silva I, De Souza N, et al. Genetic polymorphism of GSTM1 in women with breast cancer and interact with reproductive history and several clinical pathologies. Biol Res. 2005;38:273–281. doi:10.4067/s0716-97602005000200017
23. Lizard-Nacol S, Coudert B, Colosetti P, et al. Glutathione S-transferase M1 null genotype: lack of association with tumour characteristics and survival in advanced breast cancer. Breast Cancer Res. 1999;1(1):81–87. doi:10.1186/bcr17
24. Saadat I, Omidvari SH, Saadat M. Genetic polymorphism of the glutathione S-transferase M1 and development of breast cancer. Iran Biomed J. 2001;5(1):21–25.
25. Demir E. Polymorphisms Of Glutathione S-Transferase Genes (Gstm1, Gstp1, And Gstt1) And Breast Cancer Susceptibility In The Turkish Population. The Institute Of Engineering and Science Of Bilkent University; 2002:1–98.
26. Yang G, Shu X, Ruan Z, et al. Genetic polymorphisms in glutathione-S-transferase genes (GSTM1, GSTT1, GSTP1) and survival after chemotherapy for invasive breast carcinoma. Am Cancer Soc. 2004;103(1):52–58.
27. Bhattacharjee P, Paul S, Banerjee M, et al. Functional compensation of glutathione S-transferase M1 (GSTM1) null by another GST superfamily member GSTM2. Sci Rep. 2013;3:2704. 42. doi:10.1038/srep02704.
28. Andrew R, Wallis L, Harwood CH, et al. Genetic and environmental contributions to variation and population divergence in a broad-spectrum foliar defense of Eucalyptus tricarpa. Ann Bot. 2010;105:707–717. doi:10.1093/aob/mcq046
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.