Back to Journals » Therapeutics and Clinical Risk Management » Volume 12
Association of glutathione S-transferase T1, M1, and P1 polymorphisms in the breast cancer risk: a meta-analysis
Authors Song Z , Shao C, Feng C, Lu Y, Gao Y, Dong C
Received 15 January 2016
Accepted for publication 4 March 2016
Published 12 May 2016 Volume 2016:12 Pages 763—769
DOI https://doi.org/10.2147/TCRM.S104339
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Deyun Wang
Zhiwang Song,1 Chuan Shao,2 Chan Feng,1 Yonglin Lu,1 Yong Gao,1 Chunyan Dong1
1Department of Oncology, Shanghai East Hospital, Tongji University, Shanghai, 2Department of Neurosurgery, The Second Clinical Medical College of North Sichuan Medical College, Nanchong, Sichuan, People’s Republic of China
Background: Several case–control studies investigating the relationship between genetic polymorphisms of glutathione S-transferase (GST) M1, GSTT1, and GSTP1 (rs1695) and the risk of breast cancer have reported contradictory results. We therefore performed a meta-analysis to clarify this issue.
Materials and methods: An updated meta-analysis using PubMed and Web of Knowledge databases for the eligible case–control studies was performed. Random- or fixed-effects model was used.
Results: A total of 10,067 cancer cases and 12,276 controls in 41 independent case–control studies from 19 articles were included in this meta-analysis. Significant increase in risk of breast cancer for Asians was found in GSTM1-null genotype (P=0.012, odds ratio [OR] =1.17, 95% confidence interval [CI] =1.04–1.32) and GSTT1-null genotype (P=0.039, OR =1.19, 95% CI =1.01–1.41). In addition, our results showed that the GSTP1 (rs1695) polymorphisms can significantly increase the risk among Caucasians (P=0.042, OR =1.16, 95% CI =1.01–1.34). Sensitivity analysis and publication bias further confirmed the dependability of the results in this meta-analysis.
Conclusion: Our results demonstrate that both GSTM1- and GSTT1-null polymorphisms are associated with an increased risk of breast cancer in Asians and that GSTP1 Val105Ile (rs1695) polymorphism is associated with an increased breast cancer risk in Caucasians.
Keywords: GSTM1, GSTT1, GSTP1, polymorphism, breast cancer, meta-analysis
Background
Breast cancer, one of the most common cancers, has shown a steady increase in incidence worldwide in recent years. It remains the major cause of cancer-related mortality among women.1,2 According to earlier reports, there are ~1.15 million breast cancer patients diagnosed every year, and the highest incidence of breast cancer is found in Europe and USA.3,4 In the People’s Republic of China, the incidence of breast cancer has been growing rapidly. Patients with breast cancer, meanwhile, tend to be younger.5,6 Its pathogenesis is still unclear, although some studies have shown that breast cancer is caused by environmental and genetic factors.7,8
As a vital Phase II isoenzyme, the glutathione S-transferase (GST) family can identify environmentally hazardous materials and regulate the level of other enzymes and proteins in the cell. Thus, it plays an important role in many basic physiological processes of the human body.9–11 According to their distinct isoelectric points, human GSTs can be divided into seven classes, alpha (α), mu (μ), omega (ω), pi (π), sigma (σ), theta (θ), and zeta (ς). There are also microsomal GST isoenzymes.12 It is reported that there are at least three genes of them with common functional polymorphisms, which are GSTT1 (θ), GSTM1 (μ), and GSTP1 (π). Every mutation in each of them may potentially lead to a loss of enzymatic function.13,14 Many researchers have shown that GSTs are crucial to cellular protection from a great deal of damage, and the polymorphism of GSTs could result in cancers of the esophagus,15 kidney,16 and liver,17 and glioma.18
A large number of studies have indicated that the GSTT1, GSTM1, and GSTP1 (rs1695) polymorphisms are associated with breast cancer.8,11–38 However, the results of these studies are inconclusive. Therefore, we performed this meta-analysis of published case–control studies to solve the conflicting results and draw a relatively reliable conclusion.
Materials and methods
Literature search
All related studies published before May 31, 2015, were identified independently by two reviewers through a computer-based search of PubMed (www.ncbi.nlm.nih.gov/pubmed) and Web of Knowledge (http://isiknowledge.com/) databases. The search terms used in this study were as follows: (“glutathione S-transferase” OR “GST” OR “GSTT1” OR “GSTM1” OR “GSTP1”) AND (“breast cancer” OR “breast neoplasm” OR “breast carcinoma”) AND “polymorphism”. There was no language restriction. For this meta-analysis, the included studies had to meet the following criteria: 1) a case–control study on the polymorphism of GSTT1, GSTM1, or GSTP1 polymorphism and the risk of breast cancer; 2) reported genotype frequencies of cases and controls; and 3) the genotypes of control subjects in accordance with the Hardy–Weinberg equilibrium (HWE).
Data extraction
Two investigators extracted carefully the relevant information independently, and any discrepancy was settled by consensus. The following data were extracted from articles: first author’s name, year, country, ethnicity, the source of controls, and the genotype attribution of cases and controls.
Statistical analysis
The odds ratio (OR) and their 95% confidence interval (CI) were adopted to evaluate the strength of association between the polymorphism of GSTT1, GSTM1, and GSTP1 (rs1695) and the risk of breast cancer. First, we examined GSTT1 and GSTM1 genotypes using the null vs present model. Then, the relationship between the GSTP1 (rs1695) polymorphism and risk of breast cancer was estimated with allelic (V vs I) model, the recessive (VV vs II + VI), the dominant (VV + VI vs II), and the codominant (VV vs II). The statistical significance of the pooled OR was determined by the Z-test, and a P<0.05 was considered statistically significant. HWE was estimated using the chi-squared test among controls, where P<0.05 was considered a significant departure from HWE. We evaluated heterogeneity among included studies with chi-squared-based Q-test and I2 statistic. If the heterogeneity was obvious, with P<0.1, random-effects model was used to calculate the pooled OR; otherwise, the fixed-effects models were adopted. Moreover, subgroup analysis was conducted by ethnicity.
We performed sensitivity analysis by omitting single study every time to assess the robustness of the results. Funnel plots and Egger’s tests were used to explore the potential publication bias; P>0.05 was considered to indicate no significant publication bias. All P-values were based on two-sided tests.
Results
Study characteristics
Our meta-analysis was conducted according to guidelines of the “Preferred Reporting Items for Systematic Reviews and Meta-Analyses” (PRISMA) statement (checklist)39 and “Meta-analysis on Genetic Association Studies” statement (checklist).40 The flowchart is illustrated in Figure 1. A total of 171 potentially relevant articles were found by the literature search, and among these 121 articles were excluded because of obvious irrelevance after a preliminary screening of the titles and abstracts. In addition, after full-text reviews of the remaining 33 articles, 14 articles were excluded for the following reasons: articles were based on studies on prognosis or chemotherapy sensitivity (n=9), article was a quantitative analysis (n=1), article was a case report (n=1), articles had insufficient data (n=3), and studies deviated from HWE (n=2). Articles reporting data for different kinds of GST ethnicity were treated as independent studies. Finally, 19 articles8,19–36 involving 41 independent case–control studies with 10,067 cancer cases and 12,276 controls completely met the inclusion criteria. The detailed data collected from the included studies are summarized in Table 1.
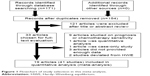  | Figure 1 Flowchart of study selection in this meta-analysis. |
  | Table 1 Characteristics of the literature included in the meta-analysis |
Association of GSTM1-null polymorphism with breast cancer risk
Seventeen studies including 4,046 cases and 5,344 controls studied the association between GSTM1-null polymorphism and breast cancer.8,19,21–26,28,30–31,33–36 Our meta-analysis showed that there was no significant association of GSTM1-null polymorphism with breast cancer risk (OR =1.13, 95% CI =0.97–1.32) (Table 2). When stratifying for ethnicity, we found that GSTM1-null polymorphism could increase the breast cancer risk for Asians (OR =1.17, 95% CI =1.04–1.32) (Figure 2). However, no significant association was found for Caucasians (OR =1.13, 95% CI =0.85–1.52) or mixed ethnicity (OR =0.90, 95% CI =0.62–1.30) (Table 2).
  | Figure 2 Forest plot for the association of GSTM1 null polymorphism and breast cancer risk for Asians. |
Association of GSTT1-null polymorphism with breast cancer risk
Fourteen studies including 2,788 cases and 3,686 controls studied the association of GSTT1-null polymorphism with breast cancer.8,19,21–23,25,28,30–31,34–36 Totally, our meta-analysis showed that there was no significant association between GSTT1-null polymorphism and the risk of breast cancer (OR =1.15, 95% CI =0.93–1.42) (Table 2). When stratifying for ethnicity, similarly, we found that GSTT1-null polymorphism could increase breast cancer risk among Asians (OR =1.19, 95% CI =1.01–1.41) (Figure 3). However, we found that there was no significant association of GSTT1-null polymorphism with breast cancer risk for Caucasians (OR =1.17, 95% CI =0.96–1.42) or mixed ethnicity (OR =0.88, 95% CI =0.57–1.34) (Table 2).
  | Figure 3 Forest plot for the association of GSTT1 null polymorphism and breast cancer risk for Caucasians. |
Association of GSTP1 Val105Ile polymorphism with breast cancer risk
Ten studies including 3,233 cases and 3,246 controls studied the association between GSTP1 Val105Ile (rs1695) polymorphism and breast cancer.20,23–24,27,29–32,34,36 In the allelic model, our meta-analysis showed that GSTP1 Val105Ile (rs1695) polymorphism was not associated with breast cancer risk overall (OR =1.21, 95% CI =0.99–1.48) (Table 2). When stratifying for ethnicity, similarly, we found that GSTP1 Val105Ile polymorphism could increase breast cancer risk for Caucasians (OR =1.16, 95% CI =1.01–1.34) (Figure 4). However, we found that there was no significant association between GSTP1 Val105Ile (rs1695) polymorphism and breast cancer risk for Asians (OR =1.26, 95% CI =0.91–1.75) (Table 2).
In the recessive model, we found that GSTP1 Val105Ile (rs1695) polymorphism was not associated with breast cancer risk overall (OR =1.16, 95% CI =0.83–1.62) (Table 2). When stratifying for ethnicity, we found that GSTP1 Val105Ile (rs1695) polymorphism had no significant association with the risk of breast cancer for Caucasians (OR =1.14, 95% CI =0.86–1.52) (Table 2) or for Asians (OR =1.28, 95% CI =0.70–2.35) (Table 2).
Similarly, we did not find any significant association of GSTP1 Val105Ile (rs1695) polymorphism with breast cancer risk overall (OR =1.19, 95% CI =0.93–1.52) and for Caucasians (OR =1.03, 95% CI =0.85–1.25) or Asians (OR =1.34, 95% CI =0.94–1.93) (Table 2) in the dominant model.
In codominant model, we found that there was no significant association of GSTP1 Val105Ile (rs1695) polymorphism with breast cancer risk overall (OR =1.24, 95% CI =0.81–1.89) and for Caucasians (OR =1.14, 95% CI =0.84–1.57) or Asians (OR =1.45, 95% CI =0.69–3.05) (Table 2).
Sensitivity analysis
A single study was excluded each time to reflect the effect of an individual study on the pooled OR and 95% CI. The deletion of any single study did not qualitatively alter the corresponding pooled ORs; these findings confirmed the stability of our meta-analysis results (data not shown).
Publication bias
We performed both Begg’s and Egger’s tests and generated a funnel plot to evaluate any potential publication bias. The symmetry of the funnel plots indicated no statistical evidence of publication bias in this meta-analysis (data not shown).
Discussion
Large-scale epidemiological studies on gene polymorphisms can contribute to uncovering the role and the corresponding mechanism of genes in the development of many diseases. To the best of our knowledge, this is the most comprehensive meta-analysis that evaluated the association of GSTM1-null, GSTT1-null, and GSTP1 Val105Ile (rs1695) polymorphisms with the risk of breast cancer. The obvious strength of meta-analysis is based on the accumulation of published data, providing a greater amount of information to find significant differences. In total, the meta-analysis involved 41 independent case–control studies of 19 articles comprising 10,067 cancer cases and 12,276 controls.
Our results demonstrate that the GSTM1-null, GSTT1-null, and GSTP1 Val105Ile (rs1695) polymorphisms are not significantly associated with breast cancer risk in the overall populations. However, in the stratified analysis by ethnicity, significant associations were found in Asians for GSTM1-null and GSTT1-null polymorphisms. Significant result was also obtained for GSTP1 Val105Ile (rs1695) polymorphism among Caucasians. However, no significant associations were found among Caucasian and mixed populations for GSTM1-null and GSTT1-null polymorphism. Similarly, no significant associations were found among Asians for GSTP1 Val105Ile (rs1695) polymorphism.
In 2013, Liu et al37 performed a meta-analysis, which showed that GSTP1 Val105Ile (rs1695) polymorphism was associated with the susceptibility of breast cancer in Asians under the allelic and recessive model. In another meta-analysis study, the GSTM1 and GSTP1 polymorphisms (under allelic and dominant model) were found to be associated with increased breast cancer risk Asian population, especially in East Asians, and that the GSTT1 polymorphism might not be associated with breast cancer.38 These differences between different meta-analyses might have been due to the relatively small number of samples in each study.
There are several possible causes for the differences between different ethnicities. First of all, the frequencies of the genotype vary sharply between different ethnicities. For instance, the homozygous null genotype distributions of the GSTT1 polymorphism change greatly between Asian and Caucasian populations, with a prevalence of 79.6% and 19.0%, respectively.23,41 Therefore, more studies with larger sample sizes are needed to further confirm ethnic difference in the association between these polymorphisms and breast cancer risk. Second, different lifestyles may explain partially the ethnic difference, as Asians and Caucasian adopt different food preferences. Previous studies have demonstrated that high intake of certain fruits, vegetables, milk, and eggs may have important effects on breast cancer risk.42–44 Different lifestyles, such as maintaining body mass index, physical exercise, and intake of sugary drinks, red meat, and alcohol, also have important influence in breast cancer susceptibility.45,46 Finally, the finding of an increasing breast cancer risk only in Asians is a chance of finding because of the relatively small number of the studies among each ethnicity included in this meta-analysis.
GSTs are important Phase II detoxification enzymes involved in the metabolism of a large number of potential carcinogens. Mutations in all of the three GST genes may lead to oxidative stress and the accumulation of reactive quinone intermediates in cells. In the GST family, it is well known that the proteins GSTM1, GSTT1, and GSTP1 (rs1695) have important influence on the modification of some vital enzymes. Many studies have shown that these enzymes may combine with glutathione and affect the detoxification of electrophilic compounds, including carcinogens, therapeutic drugs, environmental toxins, and products of oxidative stress.47,48
Limitations
Several limitations of our study should be acknowledged when interpreting the results. First, due to the failure in acquiring detailed original information, all the results of this meta-analysis is based on single-factor calculation without adjustment by other important co-variables, such as menopausal state, age of menarche, tobacco smoking habit, lifestyle factors, and family history. Second, some heterogeneity was observed in this study due to uncontrolled confounding factors and selection bias. We solved this problem by adopting a random-effects model and performing sensitivity analysis. Third, only articles published and written in English were included this meta-analysis, which might have resulted in some degree of publication bias. However, no significant publication bias was detected, indicating that no noticeable harm was done by potential publication bias.
Conclusion
Our meta-analysis demonstrates that GSTM1- and GSTT1-null polymorphisms can increase breast cancer risk for Asians, and GSTP1 Val105Ile (rs1695) polymorphism can increase breast cancer risk for Caucasians.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (81573008, 81201798), the Fund of Pudong Health Bureau of Shanghai (PWRd2014-01), and the Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (PWZxq2014-04).
Disclosure
The authors report no conflicts of interest in this work.
References
Pedraza AM, Pollán M, Pastor-Barriuso R, Cabanes A. Disparities in breast cancer mortality trends in a middle income country. Breast Cancer Res Treat. 2012;134(3):1199–1207. | ||
Hortobagyi GN, de la Garza Salazar J, Pritchard K, et al; ABREAST Investigators. The global breast cancer burden: variations in epidemiology and survival. Clin Breast Cancer. 2005;6(5):391–401. | ||
Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220–241. | ||
Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378(9801):1461–1484. | ||
Yu KD, Di GH, Wu J, et al. Development and trends of surgical modalities for breast cancer in China: a review of 16-year data. Ann Surg Oncol. 2007;14(9):2502–2509. | ||
Yang L, Parkin DM, Li L, Chen Y. Time trends in cancer mortality in China: 1987–1999. Int J Cancer. 2003;106(5):771–783. | ||
Jo YH, Kim HO, Lee J, et al. MCPH1 protein expression and polymorphisms are associated with risk of breast cancer. Gene. 2013;517(2):184–190. | ||
Luo J, Gao YT, Chow WH, et al. Urinary polyphenols, glutathione S-transferases copy number variation, and breast cancer risk: results from the Shanghai women’s health study. Mol Carcinog. 2012;51(5):379–388. | ||
Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. | ||
Atkinson HJ, Babbitt PC. Glutathione transferases are structural and functional outliers in the thioredoxin fold. Biochemistry. 2009;48(46):11108–11116. | ||
Udomsinprasert R, Pongjaroenkit S, Wongsantichon J, et al. Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme. Biochem J. 2005;388(Pt 3):763–771. | ||
Dong Y, Li C, Zhang Y, et al. Glutathione S-transferase gene family in Gossypium raimondii and G. arboreum: comparative genomic study and their expression under salt stress. Front Plant Sci. 2016;7:139. | ||
Alves S, Amorim A, Ferreira F, Norton L, Prata MJ. The GSTM1 and GSTT1 genetic polymorphisms and susceptibility to acute lymphoblastic leukemia in children from north Portugal. Leukemia. 2002;16(8):1565–1567. | ||
He HR, Zhang XX, Sun JY, et al. Glutathione S-transferase gene polymorphisms and susceptibility to chronic myeloid leukemia. Tumour Biol. 2014;35(6):6119–6125. | ||
Cai Y, Wang J. Significant association of glutathione S-transferase T1 null genotype with esophageal cancer risk: a meta-analysis. Mol Biol Rep. 2013;40(3):2397–2403. | ||
Jia CY, Liu YJ, Cong XL, et al. Association of glutathione S-transferase M1, T1, and P1 polymorphisms with renal cell carcinoma: evidence from 11 studies. Tumour Biol. 2014;35(4):3867–3873. | ||
Shen YH, Chen S, Peng YF, et al. Quantitative assessment of the effect of glutathione S-transferase genes GSTM1 and GSTT1 on hepatocellular carcinoma risk. Tumour Biol. 2014;35(5):4007–4015. | ||
Zhang B, Wang J, Niu H, et al. Association between glutathione S-transferase T1 null genotype and glioma susceptibility: a meta-analysis. Tumour Biol. 2014;35(3):2081–2086. | ||
Bailey LR, Roodi N, Verrier CS, Yee CJ, Dupont WD, Parl FF. Breast cancer and CYPIA1, GSTM1, and GSTT1 polymorphisms: evidence of a lack of association in Caucasians and African Americans. Cancer Res. 1998;58(1):65–70. | ||
Zhao MF, Lewis R, Gustafson DR, Wen WQ, Cerhan JR, Zheng W. No apparent association of GSTP1 A(313)G polymorphism with breast cancer risk among postmenopausal Iowa women. Cancer Epidemiol Biomarkers Prev. 2001;10(12):1301–1302. | ||
da Fonte de Amorim L, Rossini A, Mendonça G, et al. CYP1A1, GSTM1, and GSTT1 polymorphisms and breast cancer risk in Brazilian women. Cancer Lett. 2002;181(2):179–186. | ||
Sohail A, Kanwal N, Ali M, et al. Effects of glutathione-S-transferase polymorphisms on the risk of breast cancer: a population-based case-control study in Pakistan. Environ Toxicol Pharmacol. 2013;35(2):143–153. | ||
Zgheib NK, Shamseddine AA, Geryess E, et al. Genetic polymorphisms of CYP2E1, GST, and NAT2 enzymes are not associated with risk of breast cancer in a sample of Lebanese women. Mutat Res. 2013;747–748:40–47. | ||
Sakoda LC, Blackston CR, Xue K, et al. Glutathione S-transferase M1 and P1 polymorphisms and risk of breast cancer and fibrocystic breast conditions in Chinese women. Breast Cancer Res Treat. 2008;109(1):143–155. | ||
Chang TW, Wang SM, Guo YL, Tsai PC, Huang CJ, Huang W. Glutathione S-transferase polymorphisms associated with risk of breast cancer in southern Taiwan. Breast. 2006;15(6):754–761. | ||
Ambrosone CB, Coles BF, Freudenheim JL, Shields PG. Glutathione-S-transferase (GSTM1) genetic polymorphisms do not affect human breast cancer risk, regardless of dietary antioxidants. J Nutr. 1999;129(2S suppl):565S–568S. | ||
Saxena A, Dhillon VS, Shahid M, et al. GSTP1 methylation and polymorphism increase the risk of breast cancer and the effects of diet and lifestyle in breast cancer patients. Exp Ther Med. 2012;4(6):1097–1103. | ||
Syamala VS, Sreeja L, Syamala V, et al. Influence of germline polymorphisms of GSTT1, GSTM1, and GSTP1 in familial versus sporadic breast cancer susceptibility and survival. Fam Cancer. 2008;7(3):213–220. | ||
Khabaz MN. Polymorphism of the glutathione S-transferase P1 gene (Gst-Pi) in breast carcinoma. Pol J Pathol. 2014;65(2):141–146. | ||
Van Emburgh BO, Hu JJ, Levine EA, et al. Polymorphisms in CYP1B1, GSTM1, GSTT1 and GSTP1, and susceptibility to breast cancer. Oncol Rep. 2008;19(5):1311–1321. | ||
Jaramillo-Rangel G, Ortega-Martinez M, Cerda-Flores RM, Barrera-Saldana HA. Polymorphisms in GSTM1, GSTT1, GSTP1, and GSTM3 genes and breast cancer risk in northeastern Mexico. Genet Mol Res. 2015;14(2):6465–6471. | ||
Ge J, Tian AX, Wang QS, et al. The GSTP1 105Val allele increases breast cancer risk and aggressiveness but enhances response to cyclophosphamide chemotherapy in North China. PLoS One. 2013;8(6):e67589. | ||
Maugard CM, Charrier J, Bignon YJ. Allelic deletion at glutathione S-transferase M1 focus and its association with breast cancer susceptibility. Chem Biol Interact. 1998;111–112:365–375. | ||
Hashemi M, Eskandari-Nasab E, Fazaeli A, et al. Association between polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and breast cancer risk in a sample Iranian population. Biomark Med. 2012;6(6):797–803. | ||
Zheng W, Wen WQ, Gustafson DR, Gross M, Cerhan JR, Folsom AR. GSTM1 and GSTT1 polymorphisms and postmenopausal breast cancer risk. Breast Cancer Res Treat. 2002;74(1):9–16. | ||
Ramalhinho AC, Fonseca-Moutinho JA, Breitenfeld Granadeiro LA. Positive association of polymorphisms in estrogen biosynthesis gene, CYP19A1, and metabolism, GST, in breast cancer susceptibility. DNA Cell Biol. 2012;31(6):1100–1106. | ||
Liu JJ, Liu JL, Zhang X, Xie L, Zeng J. A meta-analysis of the association of glutathione S-transferase P1 gene polymorphism with the susceptibility of breast cancer. Mol Biol Rep. 2013;40(4):3203–3212. | ||
Tang JQ, Zhou QX, Zhao F, et al. Association of glutathione S-transferase T1, M1 and P1 polymorphisms in the breast cancer risk: a meta-analysis in Asian population. Int J Clin Exp Med. 2015;8(8):12430–12447. | ||
Moher D, Liberati A, Tetzlaff J, Altman DG, Group TP. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. | ||
Sagoo GS, Little J, Higgins JPT. Systematic reviews of genetic association studies. PLoS Med. 2009;6: e1000028. | ||
Duggan C, Ballard-Barbash R, Baumgartner RN, Baumgartner KB, Bernstein L, McTiernan A. Associations between null mutations in GSTT1 and GSTM1, the GSTP1 Ile (105)Val polymorphism, and mortality in breast cancer survivors. Springerplus. 2013;2:450. | ||
Ambrosone CB, McCann SE, Freudenheim JL, Marshall JR, Zhang YS, Shields PG. Breast cancer risk in premenopausal women is inversely associated with consumption of broccoli, a source of isothiocyanates, but is not modified by GST genotype. J Nutr. 2004;134(5):1134–1138. | ||
Bao PP, Shu XO, Zheng Y, et al. Fruit, vegetable, and animal food intake and breast cancer risk by hormone receptor status. Nutr Cancer. 2012;64(6):806–819. | ||
Si RH, Qu KP, Jiang ZB, Yang XJ, Gao P. Egg consumption and breast cancer risk: a meta-analysis. Breast Cancer. 2014;21(3):251–261. | ||
Castello A, Martin M, Ruiz A, et al; EpiGEICAM Researchers. Lower breast cancer risk among women following the world cancer research fund and american institute for cancer research lifestyle recommendations: EpiGEICAM case-control study. PLoS One. 2015;10(5):e0126096. | ||
Shin A, Sandin S, Lof M, et al. Alcohol consumption, body mass index and breast cancer risk by hormone receptor status: women’ lifestyle and health study. BMC Cancer. 2015;15:881. | ||
Nock NL, Bock C, Neslund-Dudas C, et al. Polymorphisms in glutathione S-transferase genes increase risk of prostate cancer biochemical recurrence differentially by ethnicity and disease severity. Cancer Causes Control. 2009;20(10):1915–1926. | ||
Nakazato H, Suzuki K, Matsui H, et al. Association of genetic polymorphisms of glutathione-S-transferase genes (GSTM1, GSTT1 and GSTP1) with familial prostate cancer risk in a Japanese population. Anticancer Res. 2003;23(3C):2897–2902. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


