Back to Journals » Hepatic Medicine: Evidence and Research » Volume 7
Association of cytokine gene polymorphisms with hepatitis C virus infection in a population from Rio de Janeiro, Brazil
Authors Fabrício-Silva GM, Poschetzky B, Perez R, Carneiro dos Santos R, Cavalini L, Porto LC
Received 28 May 2015
Accepted for publication 29 July 2015
Published 2 November 2015 Volume 2015:7 Pages 71—79
DOI https://doi.org/10.2147/HMER.S89447
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Gerry Lake-Bakaar
Gustavo Milson Fabrício-Silva,1 Bruno Silva Poschetzky,2 Renata de Mello Perez,2 Ronaldo Carneiro dos Santos,2 Luciana Tricai Cavalini,3 Luís Cristóvão Porto1
1Histocompatibility and Cryopreservation Laboratory, 2Gastroenterology Service, Internal Medicine Department, 3Information Technology and Healthy Education Department, Rio de Janeiro State University, Rio de Janeiro, Brazil
Background: The host immune response is an important indicator of the outcome of hepatitis C virus (HCV) infection and disease progression. The aim of this study was to explore cytokine gene polymorphisms as a candidate for susceptibility to persistent HCV infection or HCV spontaneous clearance in a population from Rio de Janeiro, Brazil.
Methods: Genetic polymorphisms in the cytokines, tumor necrosis factor-alpha (–308), transforming growth factor-beta 1 (codons 10 and 25), interleukin-10 (IL-10; –1082 and –592), IL-6 (–174), and interferon-gamma (+874) were analyzed by polymerase chain reaction sequence-specific primers in 245 patients with chronic hepatitis C (CHC), 41 spontaneous recovery (SR) patients, and 189 healthy volunteers. Further, polymorphisms in IL-28B (rs12979860, rs12980275, and rs8099917) were assessed by real-time polymerase chain reaction in all groups.
Results: The IL-28B rs12979860 CC and rs12980275 AA genotypes were significantly associated with SR of HCV infection and response to therapy, whereas the C allele of IL-6 (–174) was associated with protection to CHC. A multivariate analysis showed that the male sex and IL-28B rs12979860 CT and TT and transforming growth factor-beta 1 (codon 10) TC genotypes were factors associated with CHC. Additionally, IL-6 (–174) C allele was increased in SR patients compared with patients with CHC.
Conclusion: IL-28B polymorphisms are associated with spontaneous clearance of HCV and response to therapy in a Brazilian population. Also, IL-6 (–174) C allele is involved in SR and decreased inflammation scores.
Keywords: interleukin-28, interleukin-6, SNP, inflammation, HCV clearance
Background
Chronic hepatitis C virus (HCV) infection commonly induces immune reactive inflammation, which results in continuous liver tissue damage and progression of liver fibrosis to cirrhosis or hepatocellular carcinoma.1 However, a small portion of patients (10%–30%) mount an immune response that successfully eliminates infection.2
Several risk factors have been investigated with the aim of identifying patients who may be predisposed to a less favorable outcome. Numerous case–control, candidate gene, and allele-association studies have examined the relationship between single-nucleotide polymorphisms (SNPs) and disease course in patients with HCV infection.3
Cytokines affect the natural course of HCV infection.2 Cytokines represent a large family of molecules, which play an important role in the initiation and regulation of immune responses, and therefore, these proteins may affect HCV infection outcome.4,5 Tumor necrosis factor-alpha (TNF-α), transforming growth factor-beta 1 (TGF-β1), interleukin-10 (IL-10), IL-6, and interferon-gamma (IFN-γ) SNPs were reported to be associated with the response to natural clearance of HCV,6 the response to antiviral therapy,7 as risk factors for HCV infection,8 or progression to hepatic fibrogenesis,9 chronic liver disease,10 liver cirrhosis,11 hepatocellular carcinoma,12 or the outcome of liver transplantation.13,14
In 2009, three genome-wide association studies (GWAS) found that genetic polymorphisms within the IFN-» locus on chromosome 19 (IL-28B rs12979860) determined the outcome of pegylated-IFN-α and ribavirin (RBV) treatment responses in patients infected with genotype 1 HCV and were associated with spontaneous clearance of acute HCV infection.15 Also, in 154 Japanese patients with HCV genotype 1, another GWAS identified two SNPs, rs8099917 and rs12980275, lying between IL-28B and IL-28A that were associated with nonresponse (NR) to treatment.16
Cytokine gene polymorphisms are strongly associated with ethnicity.17 Considering the great ethnic diversity in the world, further studies with subjects from different regions and different ethnic backgrounds would provide important information to understand the interplay between host and viral factors responsible for HCV pathogenesis and the clinical course of infection.18 In this context, Brazilian population represents a unique group for a study of the association between cytokine polymorphisms and HCV infection because it is a highly genetically diverse population, composed primarily of a pool of individuals derived from Western European Caucasians, African Blacks, and South American Amerindians.19
Objectives
The aim of our study was to investigate whether a predisposition to chronic HCV infection or spontaneous HCV clearance is associated with a particular cytokine polymorphism in a Brazilian population.
Patients and methods
Patients were followed at the Gastroenterology Service at the Pedro Ernesto University Hospital of the Rio de Janeiro State University between 2004 and 2011. Chronic hepatitis C (CHC), spontaneous recovery (SR), and healthy control groups were distributed accordingly in Table 1. Self-declared Amerindians or donors with parents from Asia were excluded as their numbers were too small to be representative.
The study protocol was conducted in accordance with the ethical principles and guidance of the Helsinki Declaration and was approved by the Ethics Committee of Pedro Ernesto University Hospital of the Rio de Janeiro State University. All subjects provided written consent to participate in the study.
Patients with CHC
All patients with CHC tested positive for HCV antibodies using a second-generation enzyme-linked immunosorbent assay and tested positive for HCV RNA with polymerase chain reaction (PCR) at least three times in a 6-month follow-up. These patients were included in the study. The sequencing of the HCV NS5B region and sequence analysis were performed as previously described.20
HCV treatment
HCV treatment consisted of 180 μg/wk pegylated-IFN-α in combination with RBV dose adjusted to body weight (1,000–1,200 mg/d) for 48 weeks for patients with HCV genotype 1. Patients with HCV genotype 2 or 3 were treated with IFN + RBV for 24 weeks. Sustained virologic response (SVR) to treatment was defined using HCV viral load (VL) as a parameter at the end of treatment. Patients were declared to have treatment failure if the VL was detectable and treatment success if the VL was undetectable. Responses were defined as SVR and NR.
Histopathology
Liver biopsies were performed in patients with CHC according to the Ishak scoring system.21 Fibrosis of the liver was staged from absent to moderate (Ishak 0–3) or advanced (Ishak 4–6) in 157 biopsies. For inflammation stages, 135 biopsies were assessed and subdivided in absent to mild inflammation (Ishak 0–8) and moderate to advanced inflammation (Ishak ≥9).
SR patients
SR patients were positive for antibody by second-generation enzyme-linked immunosorbent assay (Abbott Laboratories, Abbott Park, IL, USA) and by an immunoblot assay (INNO-LIA; Innogenetics, Ghent, Belgium) associated with a lack of detection of HCV RNA in serum (Amplicor; Hoffman-La Roche Ltd, Basel, Switzerland) on at least two occasions over an interval of at least 3 months. Patients with HIV/HCV or HBV/HCV coinfections were also excluded from this group.
Cytokine genotype typing
PCR amplification of TNF-α, TGF-β1, IL-10, IL-6, IFN-γ, and an internal control, the human β-globin gene, was carried out according to the manufacturer’s recommendations (Cytokine Genotyping Tray; One Lambda, Canoga Park, CA, USA).
The IL-28B polymorphisms rs12979860, rs12980275, and rs8099917 (ABI assay C_11710096_10) were determined with the ABI TaqMan SNP genotyping assays (Thermo Fisher Scientific, Waltham, MA, USA) using an ABI 7300 real-time PCR instrument (Thermo Fisher Scientific), and the analysis was performed as allelic discrimination.
Statistical analysis
Allele and genotype frequencies were calculated and compared among individuals of different sex and ethnic groups using Epi Info Version 3.5.1 (CDC/OMS 1996; Centers for Disease Control and Prevention, Atlanta, GA, USA – http://www.cdc.gov/epiinfo/). Hardy–Weinberg equilibrium was tested using Arlequin Version 3.5. The chi-square test with Yates’ correction or Fisher’s exact test, when appropriated, was used to compare allele and genotype frequencies among groups. A P-value <0.05 was considered to be significant using Prism software Version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA – http://www.graphpad.com). After univariate analyses, multivariate analyses were performed in R platform. Linear multivariate models were obtained by backward selection, using a P-value <0.2 for removal from the model, with ethnicity and sex included in the model.
Results
Table 1 presents demographic, sex, and histopathological (fibrosis and inflammation) characteristics as well as the viral response to treatment of the 245 patients with HCV enrolled in the current study.
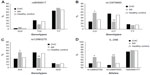
The genotype and allele frequencies of polymorphisms in IL-28B SNPs in all groups are presented in Figure 1. The frequency of the rs12979860 CC genotype was significantly higher in the SR group (58.5%) compared to the patients with CHC (26.9%) and the control (33.9%) group (P=0.0001, odds ratio [OR] =0.26, 95% confidence interval [95% CI] =0.13–0.51 and P=0.005, OR =2.75, 95% CI =1.38–5.50, respectively). Additionally, the rs12979860 CT genotype was decreased in the SR group (26.8%) compared to the CHC group (55.5%, P=0.001, OR =3.40, 95% CI =1.63–7.10) and the healthy control group (50.3%, P=0.01, OR =0.36, 95% CI =0.17–0.76).
Similarly, the frequencies of the rs12980275 AA and AG genotypes in the SR group (53.7% and 34.1%) were significantly different from the frequencies in the other groups. The AA genotype was decreased in the CHC (31.4%, P=0.009, OR =0.39, 95% CI =0.20–0.77) and the healthy control group (35.4%, P=0.04, OR =2.11, 95% CI =1.11–4.43). The AG genotype was increased only in the CHC group (53.5%, P=0.03, OR =2.22, 95% CI =1.11–4.43).
After comparing patients and controls, it was evident that the presence of the C allele at rs12979860 and the A allele at rs12980275 was strongly associated with spontaneous clearance of HCV infection. The IL-28B (rs12979860) C allele was more frequent in the SR group (72.0%) than in the CHC group (54.7%, P=0.005, OR =0.47, 95% CI =0.28–0.78) or the healthy control group (59.0%, P=0.04, OR =1.78, 95% CI =1.05–3.01). The IL-28B (rs12980275) A allele was significantly more frequent in the SR group compared to the CHC group (70.7% vs 58.2%, P=0.04, OR =0.57, 95% CI =0.34–0.95). No differences were observed in the frequency of the rs8099917 polymorphism in any of the groups.
The frequencies of additional cytokine gene polymorphisms are presented in Table 2. The frequency of the C allele SNP at IL-6 (−174) was increased in the SR group compared with the CHC group (32.9% vs 21.8%, P=0.04, OR =0.34, 95% CI =0.34–0.94). However, no differences were observed when the genotype was considered. No other significant associations were observed.
The patients’ ethnicity was also examined using self-declared data. In White subjects, the IL-28B rs8099917GG genotype was decreased in the healthy control group (2.8%) compared to the SR group (15.0%, P=0.04, OR =6.17, 95% CI =1.15–33.17). In Mestizos, the rs12979860 C allele is more frequent in the SR group (76.5%) than in the CHC group (53.9%, P=0.02, OR =0.36, 95% CI =0.15–0.82). This allele is also more frequent in the SR group in Blacks (87.5%) than in the CHC group in Blacks (50.0%, P=0.02, OR =9.25, 95% CI =1.08–79.25). No other cytokine associations were found with relation to ethnicity.
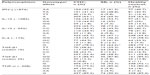
Next, the clinical characteristics of patients with chronic HCV were examined. The presence of the rs12979860 CC genotype was significantly increased in patients with SVR (P=0.001, OR =0.2, 95% CI =0.09–0.53). The IL-28B (rs12980275) AA homozygous genotype was also increased in patients with SVR (P=0.008, OR =0.30, 95% CI =0.12–0.69). Similarly, the rs12979860 C allele and the rs12980275 A allele were also increased in patients with SVR (70.3% and 73.0%, respectively) compared to NR patients (48.6%, P=0.03, OR =0.39, 95% CI =0.21–0.72 and 52.9%, P=0.007, OR =0.41, 95% CI =0.22–0.76, respectively, for the two alleles; Table 3). No other cytokine polymorphisms were associated with the SVR group.
An association between the virus genotype and a host cytokine gene polymorphism was observed only for rs8099917, where the TT genotype was more common in patients with virus genotype 1 (68.3%) than virus genotype 2 or 3 (48.6%, P=0.03, OR =2.27, 95% CI =1.11–4.65; Table 3). No other cytokine was found to influence the occurrence of viral genotypes.
No associations were observed between cytokine genotype polymorphisms and fibrosis or inflammation. With regard to allele distribution, the IL-6 (−174) C allele was significantly decreased in patients with moderate/advanced inflammation (11.5%) compared to patients with absent/mild inflammation (26.1%, P=0.04, OR =2.71, 95% CI =1.10–6.70).
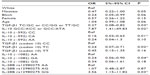
Multivariate analysis showed significant associations with parameters in the CHC group when compared to the SR group (Table 4). Male sex (P=0.0300, OR =2.20, 95% CI =1.06–4.71) and IL-28 (rs12979860) CT and TT genotypes (P<0.0001, OR =5.30, 95% CI =2.40–12.45 and P=0.0400, OR =2.95, 95% CI =1.11–8.87, respectively) were found to be associated with susceptibility to CHC. However, TGF-β1 (codon 10) TC genotype (P=0.0300, OR =0.30, 95% CI =0.09–0.85) was associated with protection to CHC infection.
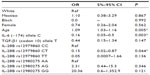
We also performed multivariate analysis to detect factors that could influence fibrosis or inflammation stage and response to treatment in patients with CHC (Tables 5 and 6). The genotypes IL-10 (−592) CA (P=0.01, OR =0.16, 95% CI =0.03–0.63), TGF-β1 (codon 10) TC (P=0.02, OR =0.24, 95% CI =0.07–0.71), and IL-10 (−1082) GA (P=0.04, OR =0.24, 95% CI =0.06–0.90) were significantly associated with stages of low fibrosis. The IL-28B rs12980275 GG genotype (P=0.03, OR =3.56, 95% CI =1.15–11.82) was associated with a higher fibrosis severity. Further, the presence of genotypes associated with intermediate cytokine production of TGF-β1 (TC/GC, CC/GG, or TT/GC; P=0.03, OR =0.27, 95% CI =0.08–0.82) confers protection to elevated grade of fibrosis, whereas IL-10 (GCC/ACC or GCC/ATA; P=0.02, OR =7.87, 95% CI =1.41–49.83) is associated with fibrosis severity (Table 5).
We also observed that the IL-6 (−174) allele C (P=0.003, OR =0.16, 95% CI =0.05–0.5) and the IL-28B rs12979860 CT genotype (P=0.04, OR =0.15, 95% CI =0.02–0.87) are protection factors to inflammatory activity. Age also had an influence on inflammation severity (P=0.005, OR =1.09, 95% CI =1.03–1.16; Table 6).
Finally, our linear multivariated model indicated some factors that were associated with response to treatment, where viral genotype 2 or 3 (P=0.015, OR =4.24, 95% CI =1.36–14.17) promotes better response to treatment, and IL-28B rs12979860 TC and TT genotypes (P=0.003, OR =0.18, 95% CI =0.05–0.53 and P=0.003, OR =0.11, 95% CI =0.02–0.45, respectively) and the intermediate producer haplotype of IL-10 GCC/ACC or GCC/ATA (P=0.008, OR =0.14, 95% CI =0.03–0.57) were associated with NR.
Discussion
Our data demonstrate the association between cytokine gene polymorphisms and the clinical course of HCV infection in a Brazilian admixed population. The most important association was observed in relation to polymorphism in IL-28. This polymorphism was significantly associated with spontaneous clearance of HCV and response to antiviral treatment.22
Currently, IL-28B has been identified to predict the response to interferon-based therapy23 with a strong association with the viral kinetics during the first days of therapy,24 natural clearance of HCV,25 and fibrosis progression.26
Thomas et al found that rs12979860 is strongly associated with spontaneous clearance of HCV spontaneously in populations of African or European ancestry, with an approximately three times higher rate of clearance in individuals with the rs12979860 genotype CC vs CT or TT.27 In the present study, we confirmed the influence of IL-28B genotypes and alleles on spontaneous clearance of HCV infection. The rs12979860 CT genotype was the most frequent in both CHC and healthy control groups (55.5% and 50.3%), but in the SR group, the most frequent genotype was CC (58.5%). Similarly, the rs12980275 AA was more frequent in the SR group (53.7%).
IL-28B polymorphisms are distributed differently among populations throughout the world.27 Brazilians are one of the most admixed populations and have a representative frequency of these SNPs. To our knowledge, we are the first group to observe an association between rs12980275 and HCV outcome in Brazil. This SNP is also associated with a sustained response to therapy in many studies.28
The risk of HCV infection varies according to ethnicity, with Asians having the highest risk.25 The observed frequency of rs12980275 in our work can be valuable for future analysis in Brazilian populations, despite this polymorphism’s strong linkage disequilibrium with rs12979860.24
IL-28B (rs8099917) TT is associated with better prognosis during interferon-based therapy. In Brazil, this association was observed by Ramos et al29 and Garcia et al30 who reported an association with spontaneous viral clearance in hepatitis C infection. However, it was not observed by Ferreira et al31 and Conde et al,32 two other works made in Brazil. Grandi et al33 in a population from south Brazil reported that the rs12979860 CC genotype was significantly associated with higher treatment response rates and a lower relapse rate compared to other genotypes. Cavalcante et al34 confirmed this result in a population from northeast Brazil, where IL-28B rs12979860 CC was associated with SVR, and the CT or TT genotypes were associated with failure of therapy. These results confirm the extreme heterogeneity in Brazil, justifying the achievement of studies in different Brazilian populations to better understand CHC outcomes.
Cavalcante et al34 also analyzed IL-28B SNPs with regard to ancestry. They observed a high (80.7%) T allele frequency that was significantly associated with SVR. In addition, the TT genotype frequency in their HCV genotype 1 population was 63%. These data are similar to our results in the CHC group (TT =65.6%), which displays one of the highest frequencies of this allele described.35 In Cavalcante’s work, the African genetic contribution determined by analyzing seven ancestry informative markers was significantly greater among patients in the nonresponder group compared with those in the SVR group. However, ancestry classified using self-reported ethnicity was not associated with HCV therapy response. We also did not find any associations with HCV infection and self-reported ethnicity in our groups, but our data suggest a role for ethnicity in the allele frequency of rs12979860 in the Mestizo and Black groups because the C allele is more frequent in the SR group (76.5% and 87.5%). The SR group also has the highest percentage of rs8099917 GG in White subjects (15.0%).
Ethnic and racial differences are known to influence HCV infection outcomes.36 Wu et al37 suggested that IL-28B allele frequency and racial descent itself can contribute to the difference in SVR rates across different population groups. Our results contribute to a better understanding of IL-28B polymorphisms and HCV infection in an admixed population like Brazilians. Self-reported ethnicity can be very useful in strategic planning aiming at an individual therapeutic approaches and adverse drug effect profile prediction in an admixed population.38 In fact, we previously reported the influence of ethnicity on the association between human leukocyte antigens and spontaneous viral clearance of HCV.39
We observed a higher frequency of the TNF-α (−308) A allele in absent/mild inflammation; further studies with a large sample should be done to confirm the relevance of this result. Recently, Talaat et al40 suggest that a TNF-α polymorphism may not be a host genetic factor associated with the severity of HCV infection but may be an independent risk factor for hepatocellular carcinoma.
In Brazil, few studies have demonstrated these associations between cytokine gene polymorphisms and HCV infection outcomes.41 Recently, Ramos et al,29 in addition to examining an association with IL-28B polymorphisms, reported the participation of IL-10 (−1082) GG and IL-4 (+3) CT genotypes during acute hepatitis C with viral clearance. We failed to find an association with these cytokines, but the controversy regarding cytokine polymorphisms and CHC42–44 encourages us to continue studying the influence of host polymorphisms and hepatitis C outcomes. Recently in a similar article, Pasha et al45 reported that IL-28B, TGF-β1 (−509), and TNF-α (−308) polymorphisms that appear to influence cytokine production may be host genetic factors associated with HCV genotype 4 infection susceptibility and response to combined antiviral treatment, demonstrating the importance of host genetic factors in HCV infection outcome.
IL-6 is a circulating cytokine secreted by activated macrophages and lymphocytes, which acts as an important modulator of immune responses.46 In HBV infection, the IL-6 (−174) CC gene polymorphism at the −174 locus affects the outcome of chronic hepatitis B.47 In HCV infection, Pavón-Castillero et al48 reported that the IL-6 serum concentration is a factor that is independently associated with SVR. We found a higher frequency of the IL-6 (−174) C allele in SR patients compared with patients with CHC, confirming the data from Barrett et al.49 This is the first reported association study of IL-6 polymorphisms and spontaneous viral clearance of hepatitis C in Brazil.
In our study, we failed to detect a direct effect of TGF-β1 on HCV infection outcome. However, multivariate analysis revealed an association between the TGF-β1 (codon 10) genotype and CHC outcome and severity of fibrosis. Many studies have reported an association between TGF-β1 and chronic hepatitis, but the results of these studies are controversial. Romani et al concluded that SNPs in the TGF-β1 gene cannot be used as prognostic factors for HCV infection in an Iranian population, despite the possibility that most of the population is susceptible to HCV infection.44 However, the functionally relevant TGF-β1 polymorphism (GG) at codon 25 may play a role in the rapid development of HCV-induced graft fibrosis, and the presence of the C allele in addition to younger donor age, female sex of the recipient, and HCV genotype other than 1b seems to protect against the development of advanced stages of fibrosis.50
In Brazil, Pereira et al identified a statistically significant relationship between the TGF-β1 codon 25 polymorphism and the HCV infection; however, there was no such association with polymorphisms in codon 10.41 Here, we have investigated the same cytokine polymorphisms, and we found an association with only codon 10 of the TGF-β1 gene rather than codon 25.
No association in IFN-γ (+874) was found, confirming recent findings in which this polymorphism seems to have any effect on the outcome of therapy in patients with HCV infection.51
A limitation of the present study was the small number of SR group, which restricts statistical power, inference, and generalizability. Therefore, our results should be interpreted with caution. Further studies in other populations or comprising a large cohort are necessary to reevaluate our findings and to establish whether our results can be replicated or indeed whether we may have missed positive associations with regard to cytokine polymorphisms.
Conclusion
In conclusion, our results demonstrated that the SNPs rs12979860 and rs12980275 upstream of IL-28B are associated with spontaneous clearance of HCV and treatment response in a population from Rio de Janeiro, Brazil. Furthermore, we found an influence of the IL-6 (−174) C allele on the clearance of HCV. Our results concerning histopathological characteristics provide important insights to better understand HCV infection outcomes. Since Brazil is a continental country, with many different genetic background, our study is useful to show some genetic characteristics in patients with CHC in Rio de Janeiro population.
Acknowledgments
This work is dedicated to Dr Fatima AF Figueiredo, deceased in 2014. She was adjunct professor at the Medical Sciences School, Rio de Janeiro State University, and was supervisor of Bruno Silva Poschetzky and Ronaldo Carneiro dos Santos, Master in Sciences diploma.
This study was partially funded with grants of LCPorto from FAPERJ (Carlos Chagas Filho Research Foundation of the Rio de Janeiro State, E-26/110.334/2010, E-26/111.695/2011, and E-26/102.826/2012) and from Brazil Research Council – CNPq (303794/2010-0) and Ministry of Education – Capes.
Disclosure
The authors report no conflicts of interest in this work.
References
El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–1273. e1261. | |
Amini M, Poustchi H. Hepatitis C virus spontaneous clearance: immunology and genetic variance. Viral Immunol. 2012;25(4):241–248. | |
Mosbruger TL, Duggal P, Goedert JJ, et al. Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus. J Infect Dis. 2010;201(9):1371–1380. | |
Thursz M, Yee L, Khakoo S. Understanding the host genetics of chronic hepatitis B and C. Semin Liver Dis. 2011;31(2):115–127. | |
Sofian M, Aghakhani A, Farazi AA, et al. Serum profile of T helper 1 and T helper 2 cytokines in hepatitis C virus infected patients. Hepat Mon. 2012;12(12):e6156. | |
Kimura T, Saito T, Yoshimura M, et al. Association of transforming growth factor-beta 1 functional polymorphisms with natural clearance of hepatitis C virus. J Infect Dis. 2006;193(10):1371–1374. | |
Dogra G, Chakravarti A, Kar P, Chawla YK. Polymorphism of tumor necrosis factor-alpha and interleukin-10 gene promoter region in chronic hepatitis C virus patients and their effect on pegylated interferon-alpha therapy response. Hum Immunol. 2011;72(10):935–939. | |
Knapp S, Hennig BJ, Frodsham AJ, et al. Interleukin-10 promoter polymorphisms and the outcome of hepatitis C virus infection. Immunogenetics. 2003;55(6):362–369. | |
Wang H, Mengsteab S, Tag CG, et al. Transforming growth factor-beta1 gene polymorphisms are associated with progression of liver fibrosis in Caucasians with chronic hepatitis C infection. World J Gastroenterol. 2005;11(13):1929–1936. | |
Ray S, Broor SL, Vaishnav Y, et al. Transforming growth factor beta in hepatitis C virus infection: in vivo and in vitro findings. J Gastroenterol Hepatol. 2003;18(4):393–403. | |
Radwan MI, Pasha HF, Mohamed RH, Hussien HI, El-Khshab MN. Influence of transforming growth factor-beta1 and tumor necrosis factor-alpha genes polymorphisms on the development of cirrhosis and hepatocellular carcinoma in chronic hepatitis C patients. Cytokine. 2012;60(1):271–276. | |
Wei YG, Liu F, Li B, et al. Interleukin-10 gene polymorphisms and hepatocellular carcinoma susceptibility: a meta-analysis. World J Gastroenterol. 2011;17(34):3941–3947. | |
Rattanasiri S, McDaniel DO, McEvoy M, et al. The association between cytokine gene polymorphisms and graft rejection in liver transplantation: a systematic review and meta-analysis. Transpl Immunol. 2013;28(1):62–70. | |
Gao QJ, Liu DW, Zhang SY, et al. Polymorphisms of some cytokines and chronic hepatitis B and C virus infection. World J Gastroenterol. 2009;15(44):5610–5619. | |
Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. | |
Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41(10):1105–1109. | |
Hoffmann SC, Stanley EM, Cox ED, et al. Ethnicity greatly influences cytokine gene polymorphism distribution. Am J Transplant. 2002;2(6):560–567. | |
Sun XR, Wu J, Shi KQ, Tang KF. Relationship between IL-10 gene -1082A/G and -592C/A polymorphisms and the risk of hepatitis C infection: a meta-analysis. J Viral Hepat. 2013;20(9):602–611. | |
Parra FC, Amado RC, Lambertucci JR, Rocha J, Antunes CM, Pena SD. Color and genomic ancestry in Brazilians. Proc Natl Acad Sci U S A. 2003;100(1):177–182. | |
Castilho MC, Martins AN, Horbach IS, et al. Association of hepatitis C virus NS5B variants with resistance to new antiviral drugs among untreated patients. Mem Inst Oswaldo Cruz. 2011;106(8):968–975. | |
Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22(6):696–699. | |
Sharafi H, Alavian SM, Behnava B, Pouryasin A, Keshvari M. The impact of IFNL4 rs12979860 polymorphism on spontaneous clearance of hepatitis C; a case-control study. Hepat Mon. 2014;14(10):e22649. | |
Moreira S, Garcia RF, Gutberlet A, et al. A straightforward genotyping of the relevant IL28B SNPs for the prediction of hepatitis C treatment outcome. J Virol Methods. 2012;184(1–2):93–97. | |
Bochud PY, Bibert S, Negro F, et al. IL28B polymorphisms predict reduction of HCV RNA from the first day of therapy in chronic hepatitis C. J Hepatol. 2011;55(5):980–988. | |
Jimenez-Sousa MA, Fernandez-Rodriguez A, Guzman-Fulgencio M, Garcia-Alvarez M, Resino S. Meta-analysis: implications of interleukin-28B polymorphisms in spontaneous and treatment-related clearance for patients with hepatitis C. BMC Med. 2013;11:6. | |
Fabris C, Falleti E, Cussigh A, et al. The interleukin 28B rs12979860 C/T polymorphism and serum cholesterol as predictors of fibrosis progression in patients with chronic hepatitis C and persistently normal transaminases. J Med Virol. 2012;84(5):747–755. | |
Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. | |
Venegas M, Villanueva RA, Gonzalez K, Brahm J. IL28B polymorphisms associated with therapy response in Chilean chronic hepatitis C patients. World J Gastroenterol. 2011;17(31):3636–3639. | |
Ramos JA, Silva R, Hoffmann L, et al. Association of IL-10, IL-4, and IL-28B gene polymorphisms with spontaneous clearance of hepatitis C virus in a population from Rio de Janeiro. BMC Res Notes. 2012;5:508. | |
Garcia RF, Moreira S, de Araújo Ramos AL, et al. Interleukin 28B-related polymorphisms: a pathway for understanding hepatitis C virus infection? World J Gastroenterol. 2013;19(42):7399–7404. | |
Ferreira PR, Santos C, Cortes R, et al. Association between IL28B gene polymorphisms and sustained virological response in patients coinfected with HCV and HIV in Brazil. J Antimicrob Chemother. 2012;67(2):509–510. | |
Conde SR, Rocha LL, Ferreira VM, et al. Absence of correlation between IL-28B gene polymorphisms and the clinical presentation of chronic hepatitis B in an Amazon Brazilian population. Dis Markers. 2014;2014:534534. | |
Grandi T, da Silva CM, Amaral KM, et al. Response to treatment in Brazilian patients with chronic hepatitis C is associated with a single-nucleotide polymorphism near the interleukin-28B gene. Mem Inst Oswaldo Cruz. 2013;108(1):48–53. | |
Cavalcante LN, Abe-Sandes K, Angelo AL, et al. IL28B polymorphisms are markers of therapy response and are influenced by genetic ancestry in chronic hepatitis C patients from an admixed population. Liver Int. 2012;32(3):476–486. | |
Rauch A, Kutalik Z, Descombes P, et al; Swiss Hepatitis C Cohort Study, Swiss HIV Cohort Study. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138(4):.e1–.e7. | |
Sarkar M, Bacchetti P, Tien P, et al. Racial/ethnic differences in spontaneous HCV clearance in HIV infected and uninfected women. Dig Dis Sci. 2012;58(5):1341–1348. | |
Wu LS, Wang H, Geng XP. Two IL28B polymorphisms are associated with the treatment response of different genotypes of hepatitis C in different racial populations: a meta-analysis. Exp Ther Med. 2012; 3(2):200–206. | |
Soares RA, Santos PC, Machado-Coelho GL, et al. CYP2C9 and VKORC1 polymorphisms are differently distributed in the Brazilian population according to self-declared ethnicity or genetic ancestry. Genet Test Mol Biomarkers. 2012;16(8):957–963. | |
de Almeida BS, Silva GM, da Silva PM, Perez Rde M, Figueiredo FA, Porto LC. Ethnicity and route of HCV infection can influence the associations of HLA with viral clearance in an ethnically heterogeneous population. J Viral Hepat. 2011;18(10):692–699. | |
Talaat RM, Esmail AA, Elwakil R, Gurgis AA, Nasr MI. Tumor necrosis factor-alpha -308G/A polymorphism and risk of hepatocellular carcinoma in hepatitis C virus-infected patients. Chin J Cancer. 2012;31(1):29–35. | |
Pereira FA, Pinheiro da Silva NN, Rodart IF, Carmo TM, Lemaire DC, Reis MG. Association of TGF-beta1 codon 25 (G915C) polymorphism with hepatitis C virus infection. J Med Virol. 2008;80(1):58–64. | |
Vidigal PG, Germer JJ, Zein NN. Polymorphisms in the interleukin-10, tumor necrosis factor-alpha, and transforming growth factor-beta1 genes in chronic hepatitis C patients treated with interferon and ribavirin. J Hepatol. 2002;36(2):271–277. | |
Minton EJ, Smillie D, Smith P, et al; Trent Hepatitis C Study Group. Clearance of hepatitis C virus is not associated with single nucleotide polymorphisms in the IL-1, -6, or -10 genes. Hum Immunol. 2005;66(2):127–132. | |
Romani S, Azimzadeh P, Mohebbi SR, et al. Investigation of transforming growth factor-beta1 gene polymorphisms among Iranian patients with chronic hepatitis C. Hepat Mon. 2011;11(11):901–906. | |
Pasha HF, Radwan MI, Hagrass HA, Tantawy EA, Emara MH. Cytokines genes polymorphisms in chronic hepatitis C: impact on susceptibility to infection and response to therapy. Cytokine. 2013;61(2):478–484. | |
Dienz O, Rincon M. The effects of IL-6 on CD4 T cell responses. Clin Immunol. 2009;130(1):27–33. | |
Fabris C, Toniutto P, Bitetto D, et al. Gene polymorphism at the interleukin 6 -174 G > C locus affects the outcome of chronic hepatitis B. J Infect. 2009;59(2):144–145. | |
Pavón-Castillero EJ, Muñoz-de-Rueda P, López-Segura R, et al. Importance of IL-10 and IL-6 during chronic hepatitis C genotype-1 treatment and their relation with IL28B. Cytokine. 2013;61(2):595–601. | |
Barrett S, Collins M, Kenny C, Ryan E, Keane CO, Crowe J. Polymorphisms in tumour necrosis factor-alpha, transforming growth factor-beta, interleukin-10, interleukin-6, interferon-gamma, and outcome of hepatitis C virus infection. J Med Virol. 2003;71(2):212–218. | |
Eurich D, Bahra M, Boas-Knoop S, et al. Transforming growth factor beta1 polymorphisms and progression of graft fibrosis after liver transplantation for hepatitis C virus – induced liver disease. Liver Transplant. 2011;17(3):279–288. | |
Sarvari J, Norozian H, Fattahi MR, Pirbonyeh N, Moattari A. The role of interferon gamma gene polymorphism (+874A/T, +2109A/G, and −183G/T) in response to treatment among hepatitis C infected patients in fars province, Southern Iran. Hepat Mon. 2014;14(1):e14476. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.