Back to Journals » Infection and Drug Resistance » Volume 13
Association of CRISPR/Cas System with the Drug Resistance in Klebsiella pneumoniae
Authors Wang G , Song G , Xu Y
Received 11 March 2020
Accepted for publication 9 June 2020
Published 23 June 2020 Volume 2020:13 Pages 1929—1935
DOI https://doi.org/10.2147/IDR.S253380
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Gang Wang,* Guobin Song,* Yuanhong Xu
Department of Clinical Laboratory, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yuanhong Xu Email [email protected]
Background: Klebsiella pneumoniae is a common opportunistic pathogen and its production of extended-spectrum β-lactamases (ESBL) and carbapenemases leads to drug resistance. Clustered regularly interspaced short palindromic repeats (CRISPRs) and their associated genes (Cas) are widespread in the genome of many bacteria and are a defense mechanism against foreign invaders such as plasmids and viruses.
Purpose: To investigate the prevalence of the CRISPR/Cas system in wild type strains of K. pneumoniae in the hospital and its association with drug resistance.
Materials and Methods: A total of 136 strains were collected and characterized their susceptibility to antimicrobial agents. The prevalence of CRISPR/Cas system was detected by PCR and DNA sequencing was analyzed by CRISPRFinder. The statistical analysis of the results was performed by SPSS.
Results: We found that 50/136 (37%) isolates produced ESBL and 30/136 (22%) isolates were resistant to carbapenems. These isolates were liable to be multidrug resistant against β-lactams, quinolones, and aminoglycosides. Among the carbapenem-resistant isolates, blaKPC was the main drug resistance-associated gene and different types of ESBL and AmpC genes were present. Resistance to β-lactams, quinolones, aminoglycosides, tetracyclines, and β-lactams/enzyme inhibitor were higher in absence of the CRISPR/Cas system. Eighteen spacers within the CRISPR arrays matched with the genomes of plasmids or phages, some of which carried drug resistance genes.
Conclusion: ESBL-producing and carbapenem-resistant K. pneumoniae are more likely to develop multidrug resistance and show an inverse correlation between drug resistance and CRISPR/Cas system. Absence of CRISPR/Cas modules allow for the acquisition of external drug resistance genes.
Keywords: CRISPR/Cas, Klebsiella pneumoniae, extended-spectrum β-lactamases, carbapenemases, drug resistance
Introduction
The evolutionary ability of bacteria to adapt to new environments has been favored by the acquisition of genes through horizontal gene transfer (HGT) mechanisms such as plasmid insertion.1 The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated proteins (Cas) modules are encoded by most archaea and many bacteria and are the adaptive defence systems against invading genetic elements such as viruses and plasmids.2 CRISPR arrays are composed of small direct repeats of 21 to 48 base pairs (bp), which are separated by similarly-sized spacer sequences ranging from 26 to 72 bp.3 Accompanying CRISPR sequences, there are 4–10 Cas genes which are highly conserved and encode the Cas proteins. The CRISPR/Cas system integrates a small piece of foreign DNA from invaders such as plasmids and viruses into their direct repeat sequences and will recognize and degrade the same external DNA elements during future invasions.4 Overall, the CRISPR/Cas system mediates immunity to invading genetic elements through a three-step process: adaptation, expression, and interference. During the adaptation stage, short pieces of DNA homologous to virus or plasmid sequences are recognized by a several nucleotides long protospacer adjacent motifs (PAMs) of the CRISPR/Cas system and are integrated into the CRISPR loci with assistance of Cas proteins. The second stage is expression, during which the long primary transcript of a CRISPR locus (pre-crRNA) is generated and processed into short crRNAs. At the third step, the foreign DNA or RNA is targeted and cleaved within the protospacer sequence under guidance of crRNAs in association with Cas protein complexes.5 For many bacteria, antibiotic resistance is mediated by acquisition of new DNA that is frequently encoded on mobile gene elements from plasmids and transposons. Studies have shown a highly significant inverse correlation between the presence of CRISPR/Cas system and acquired antibiotic resistance in Enterococcus faecalis.6 Some studies proposed the potential of CRISPR/Cas system to regulate bacterial virulence. In Pseudomonas aeruginosa, CRISPR/Cas system enable modulation of biofilm production, which is an important virulence factor for various pathogenic microorganisms.7 In Streptococcus pyogenes, the presence of CRISPR/Cas modulates prophage contents and hence its virulence.8 However, a study on Escherichia coli demonstrated that CRISPR/Cas system seems to ineffectively block the spread of plasmids and antibiotic resistance.9
K. pneumoniae has emerged as a dominant opportunistic pathogen in hospital environments due to the high rate of antibiotic resistance and high degree of dissemination.10 The multidrug resistance of this bacterium has been associated with the presence of high molecular weight plasmids.11 The aim of this study was to detect the prevalence of CRISPR/Cas in wild type strains of K. pneumoiae and analyze the correlation with the drug resistance patterns.
Materials and Methods
Bacterial Isolates
A total of 136 isolates of K. pneumoniae were randomly collected in the First Affiliated Hospital of Anhui Medical University from January to April 2019. They were isolated from sputum (n = 74, 54.41%), urine (n = 29, 21.32%), blood (n = 11, 8.09%), shunt fluids (n = 7, 5.15%), wound secreta (n = 6, 4.41%), and others specimens (n = 9, 6.62%). The sources are shown in Table 1. All bacterial isolates were incubated on Columbia sheep blood agar plates at 35°C and subjected to identification by Matrix-Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS, BioMérieux, France). The susceptibility tests of piperacillin (100μg), cefotaxime (30μg), cefoperazone/sulbactam (75μg/30μg), cefmetazole (30μg), meropenem (10μg), minocycline (30μg), tobramycin (10μg), and tigecycline (15μg) were performed on Mueller-Hinton agar plate by disc diffusion method. The automated broth microdilution methods (VITEK 2 Compact, BioMérieux, France) were used to determine the susceptibility of ampicillin/sulbactam, piperacillin/tazobactam, ceftazidime, ceftriaxone, cefepime, aztreonam, levofloxacin, amikacin, gentamicin, sulfamethoxazole-trimethoprim, cefotetan, imipenem, ertapenem, ciprofloxacin, polymyxin. The results were categorized as resistant (R), intermediate (I), and susceptible (S) according to Clinical Laboratory Standards Institute Criteria (M100-S27) and the interpretation of polymyxin and tigecycline were based on European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines.
 |
Table 1 Constituent Ratios of Departments Where the Samples Originated from |
Detection of Drug Resistance Genes
The ESBL genes (blaCTX-M, blaTEM, blaSHV), carbapenemase genes (blaKPC, blaNDM, blaIMP, blaVIM, blaSME, blaGES) and AmpC genes (blaFOX, blaDHA, blaMOX, blaCIT, blaACC, blaEBC) were screened by polymerase chain reaction (PCR) as described previously.12–14 The DNA of all bacterial isolates were extracted by boiling using 1× TE solution (10 mmol/L Tris-HCl and 1 mmol/L EDTA, Sangon, China). PCR cycle was as follows: pre-denaturation at 95°C for 5 minutes, denaturation at 94°C for 1 minute, annealing at the optimal temperature of different primers for 30 seconds, and extension at 72°C for 1 minute. Denaturation, annealing, and extension steps were repeated for 35 cycles with a final extension step at 72°C for 10 minutes. PCR products were separated and detected by gel electrophoresis and were confirmed by gene sequencing (Sangon, China) and nucleotide BLAST search on GenBank.
Detection of CRISPR/Cas System
The prevalence of CRISPR/Cas was determined by PCR as described above using the following primers (Cas1-F: 5ʹ-GCTGTTTGTCAAAGTTACCCGCGAACTC and Cas1-R: 5ʹ-GGTTTTGATCGCCTCATGAGTzCACAGTTG for Cas1; Cas3-F: 5′-TGGCCGACATTTGATTCAGC-3′ and Cas3-R: 5′-CCATGCTTAACATTCATCAC-3′ for Cas3; I-E CRISPR1-F: 5′-CAGTTCCTGCAACCTGGCCT-3′ and I-E CRISPR1-R: 5′-CTGGCAGCAGGTGATACAGC-3′ for CRISPR1; I-E* CRISPR2-F: 5′-GTAGCGAAACCCTGATCAAGCG-3′ and I-E* CRISPR2-R: 5′-GCGCTACGTTCTGGGGATG-3′ for CRISPR2; I-E* CRISPR3-F: 5ʹ-GACGCTGGTGCGATTCTTGAG-3ʹ and I-E* CRISPR3-R: 5ʹ-CGCAGTATTCCTCAACCGCCT-3ʹ for CRISPR3).15,16 PCR amplified products were then subjected to DNA sequencing (Sangon, China). The sequence of Cas genes were used for BLAST nucleotide search on GenBank. The sequence of CRISPR arrays were identified with CRISPRFinder (https://crispr.i2bc.paris-saclay.fr/Server/). This algorithm locates direct repeat sequences of 23–55 bp separated by variable sequences of 25–60 bp. Spacers from CRISPR arrays were extracted from CRISPRFinder outputs. Each of the unique spacer was then analyzed for their identity on GenBank by nucleotide BLAST search.
Statistical Analysis
Chi-squared test and Fisher’s exact test were used for the statistical analysis and P < 0.05 was considered as statistical significance. All data were analyzed using SPSS version 25.0. We categorized all isolates into CRISPR/Cas-positive or CRISPR/Cas-negative and compared the distribution of ESBL, AmpC, and carbapenemase genes as well as antimicrobial susceptibility.
Results
Antimicrobial Susceptibility Tests of K. pneumoniae Isolates
We collected K. pneumoniae from patient isolates and detected production of ESBL in 37% (50/136) isolates and carbapenem-resistance in 22% (30/136) isolates. With the exception of ampicillin, to which K. pneumoniae strains are intrinsically resistant, the ESBL-producing K. pneumoniae were susceptible to carbapenems (100%), tigecycline (100%), polymyxin (100%), cefmetazole (96%), amikacin (94%), fosfomycin (82%), minocycline (78%), tobramycin (68%), levofloxacin (66%) and cefepime (66%). The carbapenem-resistance isolates were multidrug resistant (MDR) and were only susceptible to tigecycline (100%), polymyxin (100%), minocycline (87%), and cotrimoxazole (67%).
Prevalence of CRISPR/Cas System in K. pneumoniae
Using PCR for detecting Cas1, Cas3, CRISPR1, CRISPR2, and CRISPR3, we found that all of the Cas1-positive isolates were also positive for Cas3 and had at least one CRISPR array (Figures 1–3). In total, CRISPR/Cas was detected in 29/136 isolates, of which 14 isolates were CRISPR2- and CRISPR3-positive, 13 isolates were CRISPR2-positive, and 2 isolates were CRISPR3-positive. After the DNA sequencing and analysis by CRISPRFinder, the CRISPR arrays had conserved direct repeats of 28 bp: 5ʹ-GAAACACCCCCACGCATGTGGGGAAGAC-3ʹ for CRISPR2 and 5ʹ-GTCTTCCCCACGCACGTGGGGGTGTTTC-3ʹ for CRISPR3. They were separated by different spacers ranging from 32 to 40 bp. The number of spacers for CRISPR2 arrays varied from 5 to 9 and CRISPR3 arrays contained more spacers from 9 to 14. Among those spacers, 18 unique spacers were revealed to match with the genomes of plasmids or phages (Table 2).
 |
Table 2 Sequences of CRISPR/Cas Spacers Matching Plasmids or Phages |
 |
Figure 1 Presence of (A) Cas1 and (B) Cas3 genes by PCR. M indicates the DNA marker. The numeric characters represent the sequential number of different K. pneumoniae isolates. |
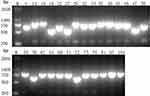 |
Figure 2 Presence of CRISPR2 gene by PCR. M indicates the DNA marker. The numeric characters represent the sequential number of different K. pneumoniae isolates which carry the CRISPR2 gene. |
 |
Figure 3 Presence of CRISPR3 gene by PCR. M indicates the DNA marker. The numeric characters represent the sequential number of different K. pneumoniae isolates which carry the CRISPR3 gene. |
Distribution of ESBL, AmpC, and Carbapenemase Genes
The distribution of drug resistance-associated genes are shown in Table 3. The majority of ESBL-producing isolates carried at least two or more ESBL genes. A single ESBL gene was found in 10 isolates, of which 6 expressed blaTEM, 2 expressed blaSHV, and 2 expressed blaCTX-M9. blaTEM was detected in 70% (35/50) of ESBL-producing isolates and 66% (33/50) expressed blaSHV, followed by CTX-M9 (48%, 24/50) and CTX-M1 (30%, 15/50). Among the 30 carbapenem-resistant isolates, the only carbapenemase gene that was detected was blaKPC. These isolates simultaneously carried ESBL and AmpC genes. One carbapenem-resistant isolate was negative for carbapenemase genes and instead harboured blaFOX, blaDHA, and blaSHV. For isolates sensitive to antimicrobial drugs, only four isolates expressed blaSHV. Taken together, blaCTX-M1, blaCTX-M9, blaTEM, blaSHV, blaKPC, blaFOX, and blaDHA were found in the 136 isolates of K. pneumoniae. The prevalence of those drug resistance-associated genes was lower in the CRISPR/Cas-positive isolates compared to those without CRISPR/Cas. Furthermore, the distribution of blaKPC, blaFOX, and blaDHA between these two groups was statistically significant (Table 4).
 |
Table 3 Genetic Characterization of 136 Isolates of K. pneumoniae |
 |
Table 4 The Distribution of Drug Resistance Genes Between CRISPR/Cas-Positive and -Negative Isolates of K. pneumoniae |
Association Between Drug Resistance and CRISPR/Cas System
The proportion of K. pneumoniae isolates resistant to antimicrobial drugs such as β-lactams, quinolones, aminoglycosides, tetracyclines, and β-lactams/enzyme inhibitors were significantly higher in the CRISPR/Cas-negative isolates than in the CRISPR/Cas-positive isolates (Table 5). CRISPR/Cas-positive isolates showed resistance to piperacillin (48.28%), aztreonam (34.48%), ceftazidime (24.14%), levofloxacin (20.69%), cefepime (13.79%), amikacin (3.45%), piperacillin/tazobactam (3.45%), and cefoperazone/sulbactam (3.45%). In comparison, the resistance rate to those drugs in CRISPR/Cas-negative isolates were of 68.22%, 55.14%, 45.79%, 41.12%, 40.19%, 24.30%, 29.91%, and 28.97%, respectively. All 29 isolates with CRISPR/Cas were susceptible to carbapenems, cefmetazole, cefotetan, and ciprofloxacin compared with the resistance rate of 28.04%, 28.97%, 26.17%, and 46.73%, respectively, in the isolates without CRISPR/Cas. ESBL production was identified in 14 of the CRISPR-positive isolates and the remaining CRISPR-positive isolates were susceptible to antimicrobial drugs.
 |
Table 5 Drug Resistance Pattern Between CRISPR/Cas-Positive and -Negative Isolates of K. pneumoniae |
Discussion
A recent bioinformatic study demonstrated that only 6/52 K. pneumoniae strains with accessible complete or draft genomes were equipped with a complete CRISPR/Cas system, which indicated CRISPR/Cas is not widely distributed in Klebsiella pneumoniae.3 We found CRISPR/Cas in 29/136 (21.3%) K. pneumoniae isolates from patient material in the hospital and the prevalence of CRISPR/Cas system was reported to be different varying from 30.7% (54/176) to 12.4% (27/217).16,17
The number, sequence, and length of CRISPR arrays and the number and sequence of the Cas proteins are highly variable between bacterial species.2 Direct repeats can be both added and deleted so that CRISPR can evolve while maintaining an identical number of repeats.18,19 The PCR results confirmed that the CRISPR arrays were at different lengths among different isolates due to the integration of spacers and the altering number of repeat-spacer units. According to a new CRISPR/Cas classification, CRISPR/Cas systems are divided into two classes and six types (class 1, including type I, III, IV and class 2, including types II, V and VI).2 The classical type I CRISPR/Cas is prevalent in bacteria, which can be divided into several subtypes. Several studies demonstrated that CRISPR/Cas systems in K. pneumoniae are classified as type I-E and I-E*.3,20 In addition to highly conserved Cas1 and Cas2, Cas3 functions as an indispensable part of type I CRISPR/Cas and has separate helicase and DNase activities.21 Both type I-E and I-E* share eight common Cas genes comprising of Cas1, Cas2, Cas3, Cas5, Cas6, Cas7, Cse1, and Cse2. Type I-E Cas3 gene location is downstream of Cas7 – Cas5, which moves upstream of Cas7 – Cas5 in type I-E*.22 In our study, we detected that K. pneumoniae isolates possess the CRISPR/Cas system based on the PCR data where Cas1 and Cas3 are present in conjunction with CRISPR arrays. Among our 136 isolates, Cas1 and Cas3 were co-expressed in 29 isolates, of which 27 isolates were CRISPR2-positive and 16 isolates were CRISPR3-positive. As described previously, the 29 isolates were classified into subtype I-E* CRISPR/Cas system.15
We compared the resistance rate of K. pneumoniae to different antimicrobial drugs based on the presence of CRISPR/Cas. Our data showed that isolates carrying this system had a lower resistance to β-lactams, quinolones, aminoglycosides, tetracyclines, and β-lactams/enzyme inhibitors. This suggests an inverse correlation between the presence of CRISPR/Cas and antibiotic resistance in wild type strains of K. pneumoniae. All the carbapenem-resistant isolates harbored blaKPC gene and AmpC or ESBL genes, which suggests that these were prone to be multidrug resistant on a genetic level. Based on the detection of seven drug resistance-associated genes, we found no statistical difference of the distribution of ESBL genes and the CRISPR/Cas system. However, blaKPC, blaFOX, and blaDHA were not detected in CRISPR/Cas-positive isolates, whether CRISPR/Cas was present significantly affected the distribution of AmpC and carbapenem genes. A previous study revealed that deletion of CRISPR/Cas cassette in K. pneumoniae increased the transformation success of blaKPC plasmids, indicating that CRISPR/Cas system mediated resistance to blaKPC plasmid invasion.15
A study revealed that decreased numbers of plasmids, phages and acquired drug resistance genes in the presence of CRISPR/Cas within the genomes of 97 K. pneumoniae. The spacers hit plasmid and phage were identified as matching with the genes of phage-related proteins.17 In our study, most of spacers matched the K. pneumoniae chromosomes and 18 unique spacers were identified matching the identity of plasmids or phages, which indicated the immunity of CRISPR/Cas system against mobile genetic elements. In addition, some of those matched foreign DNA elements that carry drug resistance genes such as blaKPC. However, it was similar to the reported finding that no plasmid- or phage-targeted spacers were identified as matching any drug resistance genes. Moreover, the two spacers (5ʹ- CCGCCGTTTAATCGCGGTGATGATATCCGGCA-3ʹ; 5ʹ-TGCCGGATATCATCACCGCGATTAAACGGCGG-3ʹ) that exist within the 25 CRISPR/Cas-positive strains matched different plasmids, which carry the genes mediating resistance to carbapenems and cephalosporins. These results demonstrated that the CRISPR/Cas system may interfere with the acquired drug resistance of K. pneumoniae to carbapenems and cephalosporins, especially for multidrug-resistant strains, which have more mobile elements linked to drug resistance genes like integrons.23 Further studies are needed to explore the interaction of CRISPR/Cas with mobile genetic elements related to drug resistance. Above all, the CRISPR/Cas system may have a very specific function to interfere with the horizontal gene transfer of drug resistance genes in K. pneumoniae.
Conclusion
The CRISPR/Cas is an adaptive defense system against foreign genetic elements and is not widely distributed in K. pneumoniae. The presence of subtype I-E* CRISPR/Cas system is associated with lower drug resistance. The spacers is a evidence that foreign genetic elements are targeted by the CRISPR/Cas system, which implies that the CRISPR arrays to some extent may prevent the acquisition of drug resistance genes in K. pneumoniae.
Ethical Approval
There is no ethical concern in this study, which was approved by The Medical Ethics Committee of the First Affiliated Hospital of Anhui Medical University. The written informed consent was obtained from patients in accordance with the Declaration of Helsinki.
Acknowledgments
This work was financially supported by the Anhui Natural Science Foundation (grant number: 9021138205).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3(9):722–732. doi:10.1038/nrmicro1235
2. Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. doi:10.1038/nrmicro2577
3. Ostria-Hernandez ML, Sanchez-Vallejo CJ, Ibarra JA, Castro-Escarpulli G. Survey of clustered regularly interspaced short palindromic repeats and their associated Cas proteins (CRISPR/Cas) systems in multiple sequenced strains of Klebsiella pneumoniae. BMC Res Notes. 2015;8:332. doi:10.1186/s13104-015-1285-7
4. Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi:10.1186/1745-6150-1-7
5. Garneau JE, Dupuis ME, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. doi:10.1038/nature09523
6. Palmer KL, Gilmore MS, Losick R. Multidrug-resistant enterococci lack CRISPR-cas. mBio. 2010;1(4):e00227–e00310. doi:10.1128/mBio.00227-10
7. Hatoum-Aslan A, Marraffini LA. Impact of CRISPR immunity on the emergence and virulence of bacterial pathogens. Curr Opin Microbiol. 2014;17:82–90. doi:10.1016/j.mib.2013.12.001
8. Louwen R, Staals RH, Endtz HP, van Baarlen P, van der Oost J. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol Mol Biol Rev. 2014;78(1):74–88. doi:10.1128/MMBR.00039-13
9. Touchon M, Charpentier S, Pognard D, et al. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology+. 2012;158(12):2997–3004.
10. El Fertas-Aissani R, Messai Y, Alouache S, Bakour R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol. 2013;61(5):209–216. doi:10.1016/j.patbio.2012.10.004
11. Garcia-Fernandez A, Villa L, Carta C, et al. Klebsiella pneumoniae ST258 producing KPC-3 identified in italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother. 2012;56(4):2143–2145. doi:10.1128/AAC.05308-11
12. Endimiani A, Carias LL, Hujer AM, et al. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae isolates possessing blaKPC in the United States. Antimicrob Agents Chemother. 2008;52(7):2680–2682. doi:10.1128/AAC.00158-08
13. Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC -lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. doi:10.1128/JCM.40.6.2153-2162.2002
14. Yu Y, Ji S, Chen Y, et al. Resistance of strains producing extended-spectrum β-lactamases and genotype distribution in China. J Infect. 2007;54(1):53–57. doi:10.1016/j.jinf.2006.01.014
15. Mackow NA, Shen J, Adnan M, Khan AS, Fries BC, Diago-Navarro E. CRISPR-Cas influences the acquisition of antibiotic resistance in Klebsiella pneumoniae. PLoS One. 2019;14(11):e225131. doi:10.1371/journal.pone.0225131
16. Lin T, Pan Y, Hsieh P, Hsu C, Wu M, Wang J. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Sci Rep. 2016;6(1).
17. Li H, Kao C, Lin W, et al. Characterization of CRISPR-Cas systems in clinical Klebsiella pneumoniae isolates uncovers its potential association with antibiotic susceptibility. Front Microbiol. 2018;9:1595.
18. Kuno S, Yoshida T, Kaneko T, Sako Y. Intricate interactions between the bloom-forming cyanobacterium microcystis aeruginosa and foreign genetic elements, revealed by diversified clustered regularly interspaced short palindromic repeat (CRISPR) signatures. Appl Environ Microbiol. 2012;78(15):5353–5360. doi:10.1128/AEM.00626-12
19. Delaney NF, Balenger S, Bonneaud C, et al. Correction: ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen. PLoS Genet. 2012;8:3. doi:10.1371/annotation/b5608bc6-aa54-40a7-b246-51fa7bc4a9db
20. Shen J, Lv L, Wang X, Xiu Z, Chen G. Comparative analysis of CRISPR-Cas systems in Klebsiella genomes. J Basic Microbiol. 2017;57(4):325–336. doi:10.1002/jobm.201600589
21. Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30(7):1335–1342. doi:10.1038/emboj.2011.41
22. Tang Y, Fu P, Zhou Y, et al. Absence of the type I-E CRISPR-Cas system in Klebsiella pneumoniae clonal complex 258 is associated with dissemination of IncF epidemic resistance plasmids in this clonal complex. J Antimicrob Chemother. 2020;75(4):890–895. doi:10.1093/jac/dkz538
23. Wei DD, Wan LG, Yu Y, et al. Characterization of extended-spectrum beta-lactamase, carbapenemase, and plasmid quinolone determinants in Klebsiella pneumoniae isolates carrying distinct types of 16S rRNA methylase genes, and their association with mobile genetic elements. Microb Drug Resist. 2015;21(2):186–193. doi:10.1089/mdr.2014.0073
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
