Back to Journals » OncoTargets and Therapy » Volume 9
Association between two interleukin-2 gene polymorphisms and cancer susceptibility: a meta-analysis
Authors Zhang M , Tan X, Huang J , Xie L, Wang H, Shi J, Lu W, Lv Z, Mei H, Liang C
Received 19 August 2015
Accepted for publication 21 January 2016
Published 19 April 2016 Volume 2016:9 Pages 2181—2192
DOI https://doi.org/10.2147/OTT.S94761
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Jianmin Xu
Meng Zhang,1,2,* Xiuxiu Tan,1,* Junjie Huang,3,* Lijuan Xie,1 Hao Wang,1 Jizhou Shi,4 Wei Lu,1 Zhaojie Lv,1 Hongbing Mei,1 Chaozhao Liang2
1Department of Urology, Shenzhen Second People’s Hospital, Clinical Medicine of Anhui Medical University, 2Department of Urology, the First Affiliated Hospital of Anhui Medical University, Hefei, 3Department of Hematology, Shenzhen Second People’s Hospital, the First Affiliated Hospital of Shenzhen University, Shenzhen, 4Department of Urology, Shengli Oilfield Central Hospital, Dongying, Peoples Republic of China
*These authors contributed equally to this work
Background: Several epidemiological studies have illustrated that polymorphisms in interleukin-2 (IL-2) were associated with diverse cancer types. However, recently published statistics were inconsistent and inconclusive. Therefore, the current meta-analysis was performed to elaborate the effects of IL-2 polymorphisms (rs2069762 and rs2069763) on cancer susceptibility.
Material and methods: A total of 5,601 cancer cases and 7,809 controls from 21 published case–control studies were enrolled in our meta-analysis. Odds ratios (ORs) with 95% confidence intervals (CIs) were estimated to assess the association between IL-2 polymorphisms and cancer susceptibility.
Results: Our study demonstrated an increased susceptibility to cancer in rs2069762 (G vs T: OR =1.268, 95% CI =1.113–1.445; GG vs TT: OR =1.801, 95% CI =1.289–2.516; GT vs TT: OR =1.250, 95% CI =1.061–1.473; GG + GT vs TT: OR =1.329, 95% CI =1.118–1.579; GG vs GT + TT: OR =1.536, 95% CI =1.162–2.030). In the subgroup analysis, increased susceptibility to cancer was identified in the hospital-based group and PHWE<0.05 (P-value of the Hardy–Weinberg equilibrium [HWE]) group. In addition, a positive association with cancer susceptibility was observed among both Chinese and non-Chinese. However, no relationship was detected between the rs2069763 polymorphism of IL-2 and cancer susceptibility.
Conclusion: To conclude, rs2069762 polymorphism of IL-2 contributed to an increased susceptibility to cancer, whereas no association was identified between rs2069763 polymorphism and cancer susceptibility. Further detailed studies are warranted to confirm our findings.
Keywords: IL-2, polymorphism, cancer, meta-analysis
Introduction
Based on available epidemiological statistics, cancer has been one of the most common causes of morbidity and mortality around the world.1 In the People’s Republic of China, the overall cancer incidence rate and death rate were 235 and 114.3 (per 100,000 population) in 2013, respectively.2 In the US, cancer is the second most common cause of death, and a total of 1,658,370 new cancer cases and 589,430 cancer deaths were expected to occur in 2015.3 Cancer is a heterogeneous disease due to the involvement of complicated risk factors. There is an increase in the evidence that implies that predisposition to cancer is related to cytokines,4 such as interleukin (IL).5 Studies suggested that accumulation of genetic variants may be implicated in carcinogenesis.6 Therefore, it would be of great importance to identify candidates for prevention and treatment of cancer.
IL-2 is an immunoregulatory cytokine produced by T helper type 1 cells when they are stimulated by mitogens, antigens, or major histocompatibility complexes on antigen-presenting cells.7 It is involved in the chemical activity of T-cell-assisted immune responses and the enhancement of natural killer cell cytolytic process as a growth factor.8–10 Pleiotropic reactions in the immune system occur when IL-2 acts as a protein, which regulates the pro- and anti-inflammatory processes.11,12 Medical scientists currently report that polymorphisms of IL-2 gene have been associated with susceptibility to a range of inflammation malfunctions and cancer,13 including gastric atrophy,14 rheumatoid arthritis,15 head and neck cancer,16 gastric cancer (GC),17 nasopharyngeal carcinoma,18 non-Hodgkin lymphoma,8 myelogenous leukemia,19 hepatocellular carcinoma (HCC),20 esophageal squamous cell carcinoma,21 breast cancer,6 and bladder cancer.22 Clinical studies had revealed that both specific and nonspecific antitumor immune responses can be augmented by transfecting IL-2 gene into tumor cells.23,24
Human IL-2 gene is encoded on chromosome 4q26 with well-characterized single-nucleotide polymorphisms, among which one (−330T/G, rs2069762) has been identified in the promoter region15 and another (+114T/G, rs2069763) at position 114 from the initiation codon in the first exon.25 Studies on IL-2 gene polymorphisms demonstrated that they play significant roles in regulating the rate of inducible expression and secretion of IL-2.26,27 The association between single-nucleotide polymorphisms of IL-2 gene and susceptibility to inflammation-based cancers, such as HCC and GC, has been reported in previously published case–control studies.7,8,10,13,17,19,28–33 However, the findings were inconsistent and inconclusive because of the limited sample size. A quantitative synthesis to accumulate all currently available statistics from various studies may uncover evidences on the relationship between genetic polymorphisms and cancer susceptibility.
Although a meta-analysis of IL-2 rs2069762 polymorphism and cancer risk has already been published recently,34 there existed some drawbacks in that study. First, only one polymorphism of IL-2 was identified, although other polymorphisms may also contribute to cancer susceptibility. Second, only ten publications were enrolled and one of them was ineligible.35 The cases and controls of these studies were limited (3,060 cases and 3,435 controls). Third, it drew a conclusion that Hardy–Weinberg equilibrium (HWE) status did not affect the relationship of IL-2 rs2069762 polymorphism and cancer susceptibility, which may not be reliable. Fourth, we found that there existed publication bias among included publications after careful calculation. To avoid the abovementioned limitations, we conducted the present meta-analysis aiming to further evaluate the association of IL-2 rs2069763 and rs2069762 polymorphisms with overall cancer susceptibility in 12 publications with 1,556 cases/2,405 controls and 4,054 cases/5,405 controls, respectively.
Materials and methods
Search strategy
We conducted a comprehensive collection research by retrieving PubMed, Web of Science, and Google Scholar databases. The keywords of retrieve were (“IL-2” OR “interleukin-2”) AND (“polymorphism” OR “variant” OR “mutation”) in combination with (“cancer” OR “tumor” OR “carcinoma” OR “leukemia”) without language restriction. The research design was limited to humans. All eligible studies were inspected carefully.
Inclusion and exclusion criteria
Only those articles satisfying the following criteria were included: 1) studies that assessed the association between IL-2 polymorphisms and cancer susceptibility; 2) studies that were case–control or cohort in design; and 3) studies from which detailed genotype frequencies of cases and controls could be obtained directly or calculated from the available data. We excluded studies when they were 1) not case–control study, such as reviews, case reports, and comments; 2) articles without sufficient data of IL-2 genotype; and 3) duplicate data.
Quality assessment
The quality of the enrolled studies was examined independently by Xiuxiu Tan and Junjie Huang referring to the Newcastle–Ottawa scale,36 which evaluates the quality of nonrandomized studies by the selection of participants, comparability of groups, and exposure assessment. Disagreement was settled as described previously.
Data extraction
The details of each satisfied study were carefully filtered by three independent investigators (Meng Zhang, Xiuxiu Tan, and Junjie Huang). Any disagreements were resolved by discussion until a consensus was reached. The name of first author, the year of publication, ethnicity, source of control, genotyping method, the number of cases and controls, and the P-value of HWE in control groups were collected from each study.
Statistical analysis
The STATA 12.0 software program (Stata Corp, College Station, TX, USA) was used to perform this meta-analysis. The odds ratio (OR) and 95% confidence interval (CI) were calculated to assess the association between IL-2 gene polymorphisms and cancer susceptibility. Five different ORs were used to compute: allele contrast model (G vs T), dominant model (GG + GT vs TT), recessive model (GG vs GT + TT), heterozygote comparison (GT vs TT), and homozygote comparison (GG vs TT) (TT, homozygotes for the common allele; GT, heterozygotes; GG, homozygotes). We adopted chi-square test-based Q statistic test to assess the heterogeneity within the case–control studies. When the Q-test (P>0.1) shows homogeneity across studies, we should choose the fixed effects model;37 otherwise, the random effects model should be selected.38 In addition, the effect of heterogeneity was quantified by the I2 value (I2<25%: no heterogeneity; I2=25%–50%: moderate heterogeneity; I2=50%–75%: high heterogeneity; and I2>75%: extreme high heterogeneity).39 We also measured the HWE of control groups. We applied stratification analyses on cancer type and ethnicity. Sensibility analysis was performed to assess the stability of the results by deleting one single case–control study each time from enrolled pooled data. In the end, the potential publication bias was evaluated by Begg’s funnel plot and Egger’s regression test. P<0.05 was considered as statistically significant.
Result
Included studies’ identification and characteristics
As shown in Figure 1, the literature research identified a total of 354 related publications. After reading the title and abstract, we reserved 18 articles concerning the association between IL-2 polymorphisms and cancer susceptibility. Additional six publications were excluded since there were no data for IL-2 polymorphisms or were duplicates or were about other IL polymorphisms. Finally, a total of 12 publications were enrolled. For IL-2 rs2069763 polymorphism, a total of five publications with six case–control studies comprising 1,556 cases and 2,405 controls were enrolled, while 12 publications with 15 case–control studies comprising 4,045 cases and 5,404 controls were included for rs2069762 polymorphism.
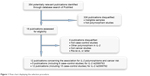  | Figure 1 Flow chart displaying the selection procedure. |
As shown in Table 1, for the six studies of IL-2 rs2069763 polymorphism, three were hospital-based designs and the others were population-based designs. In addition, three studies conformed to HWE while others did not.31,32 As for IL-2 rs2069762 polymorphism, four studies were population based and eleven were hospital based. Six conformed to HWE while others did not.7,8,13,17,28,30,31 When sorted by cancer type, there were two HCC studies, five GC studies, and eight other studies. In terms of the ethnic populations in these studies, nine were from the People’s Republic of China and six were non-Chinese.
Results of pooled meta-analysis
Tables 2 and 3 presented the results of meta-analysis, it demonstrated that no significant association between IL-2 rs2069763 and cancer susceptibility was identified (Table 2, homozygous: OR =1.039, 95% CI =0.862–1.252; heterozygous: OR =1.046, 95% CI =0.888–1.232; recessive: 0.972, 95% CI =0.835–1.133; dominant: OR =1.060, 95% CI =0.912–1.230; and allele comparing: OR =0.983, 95% CI =0.895–1.079), whereas an increased susceptibility between cancer and rs2069762 polymorphism was uncovered (Table 3, G vs T: OR =1.268, 95% CI =1.113–1.445; GG vs TT: OR =1.801, 95% CI =1.289–2.516, Figure 2; GT vs TT: OR =1.250, 95% CI =1.061–1.473; GG + GT vs TT: OR =1.329, 95% CI =1.118–1.579; GG vs GT + TT: OR =1.536, 95% CI =1.162–2.030).
Result of subgroup
Tables 2 and 3 also show the outcomes of subgroup analysis. The data in Table 2 suggested that rs2069763 polymorphism of IL-2 has no significant association with cancer susceptibility in the subgroups sorted by either source of controls or PHWE. However, as for rs2069762 polymorphism (Table 3), it demonstrated an increased susceptibility to cancer in the hospital-based studies (G vs T: OR =1.286, 95% CI =1.095–1.511; GG vs TT: OR =1.950, 95% CI =1.279–2.971; GT vs TT: OR =1.220, 95% CI =1.017–1.464; GG + GT vs TT: OR =1.310, 95% CI =1.074–1.599, Figure 3; GG vs GT + TT: OR =1.683, 95% CI =1.180–2.400) and the PHWE<0.5 group (G vs T: OR =1.341, 95% CI =1.151–1.561 and GG vs TT: OR =2.156, 95% CI =1.326–3.507, Figure 4; GT vs TT: OR =1.337, 95% CI =1.109–1.613; GG + GT vs TT: OR =1.421, 95% CI =1.175–1.720; and GG vs GT + TT: OR =1.827, 95% CI =1.173–2.845). In the subgroup meta-analysis by ethnicity, the rs2069762 polymorphism was observed with positive association with cancer susceptibility among both Chinese and non-Chinese. Specifically, the analysis indicated that non-Chinese population suffered more risk from this polymorphism (G vs T: OR =1.652, 95% CI =1.302–2.097; GG vs TT: OR =4.264, 95% CI =1.506–12.069, Figure 5; GT vs TT: OR =1.525, 95% CI =1.089–2.136; GG + GT vs TT: OR =1.647, 95% CI =1.252–2.166; and GG vs GT + TT: OR =3.341, 95% CI =1.218–9.161) than Chinese (G vs T: OR =1.162, 95% CI =1.013–1.332; GG vs TT: OR =1.438, 95% CI =1.058–1.954, Figure 5; GT vs TT: OR =1.180, 95% CI =0.982–1.418; GG + GT vs TT: OR =1.223, 95% CI =1.003–1.490; and GG vs GT + TT: OR =1.302, 95% CI =1.040–1.629). However, no relationship was identified between the rs2069762 polymorphism of IL-2 and cancer susceptibility within a certain type of cancer.
Sensitivity analysis and publication bias
We executed a sensitivity analysis to assess the influence of separate study on the pooled ORs by excluding one single study each time and a negative result was achieved (Figure 6). We detected publication bias through Begg’s funnel plot and Egger’s test. As shown in Figure 7, significant publications bias was revealed for rs2069762 (Egger’s test P=0.005), while no obvious bias was identified for rs2069763 (Egger’s test P=0.146). Therefore, trim and fill method was conducted for rs2069762 to further evaluate the publication bias. As shown in Figure 8, four theoretical studies were added and no significant difference was obtained (P<0.05), proving the stability of our results. The quality of enrolled studies is presented in Table 4.
Discussion
The association between IL-2 gene polymorphisms and susceptibility to various cancers are widely discussed these days. Unfortunately, the results are inconsistent and inconclusive. A recent meta-analysis concerning the association between rs2069762 polymorphism of IL-2 and cancer susceptibility was published, and their results suggested that IL-2 rs2069762 polymorphism was significantly associated with cancer risk. However, several limitations should be noted for this study. First, the investigators only enrolled ten publications for rs2069762 polymorphism, including ten case–controls comprising of 3,060 cases and 3,435 controls.34 However, one unqualified publication was enrolled,35 which adopted samples instead of blood tissues, distinguishing it from other enrolled studies and potentially contributing to bias. In the present work, we excluded this publication and added another three publications. We detected a total of 12 publications, including 15 case–controls comprising of 4,045 cases and 5,404 controls. Second, we also enrolled another IL-2 polymorphism (rs2069763), with five publications containing six case–control studies comprising of 1,556 cases and 2,405 controls, which broadened the scope of analysis. Third, the subgroup analysis sorted by PHWE in previous meta-analysis showed that PHWE status did not affect the correlation between IL-2-330T/G polymorphism and cancer susceptibility. Nevertheless, in our study, a significant association was found between PHWE<0.05 group and cancer susceptibility, while no association was identified between PHWE≥0.05 group and cancer susceptibility. Our work demonstrated that the PHWE status had a great effect on the pooled ORs. Although no publication bias was found in the previous study, when we excluded the ineligible study from the pooled data and assessed again, significant publication bias was uncovered.35
Although we have conducted a comprehensive retrieve and revised the disadvantages of the previous study, there are still several limitations that should be noted. First, since cancer is considered a multifactorial disease with interactions between multiple environmental exposures and individual genetic backgrounds, we failed to analyze the gene–gene and gene–environment effects in this study due to insufficient data. Second, most of the included studies were conducted in the Asian and Caucasian populations, thus, the conclusions may barely adapt to these populations. Further studies within different ethnic populations such as Africans and Latinos are warranted. Last but not least, the limited number of studies included in the meta-analysis may result in low statistical power to obtain an ideal precision of the pooled estimates.
Conclusion
In conclusion, our meta-analysis suggested that there is no association between rs2069763 polymorphism of IL-2 and cancer susceptibility, whereas an increased susceptibility to cancer was uncovered for rs2069762 polymorphism. Further well-designed studies are still warranted to further exclude the influence of PHWE<0.05 and hospital-based groups on cancer susceptibility.
Disclosure
The authors report no conflicts of interest in this work.
References
Kimman M, Norman R, Jan S, Kingston D, Woodward M. The burden of cancer in member countries of the Association of Southeast Asian Nations (ASEAN). Asian Pac J Cancer Prev. 2012;13(2):411–420. | ||
China TSCIOotPsRo [homepage on the Internet]. The Nutrition and Chronic Disease Report of 2015. 2015. Available from: www.scio.gov.cn. Accessed February 19, 2016. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. | ||
Akhurst RJ, Hata A. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov. 2012;11(10):790–811. | ||
Wang K, Grivennikov SI, Karin M. Implications of anti-cytokine therapy in colorectal cancer and autoimmune diseases. Ann Rheum Dis. 2013;72(suppl 2):ii100–ii103. | ||
Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M. Genetic susceptibility to breast cancer. Mol Oncol. 2010;4(3):174–191. | ||
Shen Y, Liu Y, Liu S, Zhang A. The association between-330T/G polymorphism of interleukin 2 gene and bladder cancer. DNA Cell Biol. 2012;31(6):983–987. | ||
Song H, Chen L, Cha Z, Bai J. Interleukin 2 gene polymorphisms are associated with non-Hodgkin lymphoma. DNA Cell Biol. 2012;31(7):1279–1284. | ||
Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775–787. | ||
Wei YS, Lan Y, Zhang L, Wang JC. Association of the interleukin-2 polymorphisms with interleukin-2 serum levels and risk of nasopharyngeal carcinoma. DNA Cell Biol. 2010;29(7):363–368. | ||
Crabtree GR, Clipstone NA. Signal transmission between the plasma membrane and nucleus of T lymphocytes. Annu Rev Biochem. 1994;63(1):1045–1083. | ||
Pyo C-W, Hur S-S, Kim Y-K, et al. Polymorphisms of IL-1B, IL-1RN, IL-2, IL-4, IL-6, IL-10, and IFN-γ genes in the Korean population. Hum Immunol. 2003;64(10):979–989. | ||
Amirzargar AA, Bagheri M, Ghavamzadeh A, et al. Cytokine gene polymorphism in Iranian patients with chronic myelogenous leukaemia. Int J Immunogenet. 2005;32(3):167–171. | ||
Pawlik A, Kurzawski M, Florczak M, Gawronska Szklarz B, Herczynska M. IL1beta+3953 exon 5 and IL-2 −330 promoter polymorphisms in patients with rheumatoid arthritis. Clin Exp Rheumatol. 2005;23(2):159–164. | ||
Togawa S, Joh T, Itoh M, et al. Interleukin-2 gene polymorphisms associated with increased risk of gastric atrophy from Helicobacter pylori infection. Helicobacter. 2005;10(3):172–178. | ||
López R, Zago MA, Eluf-Neto J, et al. Education, tobacco smoking, alcohol consumption, and IL-2 and IL-6 gene polymorphisms in the survival of head and neck cancer. Braz J Med Biol Res. 2011;44(10):1006–1012. | ||
Shin WG, Jang JS, Kim HS, et al. Polymorphisms of interleukin-1 and interleukin-2 genes in patients with gastric cancer in Korea. J Gastroenterol Hepatol. 2008;23(10):1567–1573. | ||
Seder RA. Acquisition of lymphokine-producing phenotype by CD4+ T cells. J Allergy ClinImmunol. 1994;94(6):1195–1202. | ||
Wu J, Lu Y, Ding YB, et al. Promoter polymorphisms of IL2, IL4, and risk of gastric cancer in a high-risk Chinese population. Mol Carcinog. 2009;48(7):626–632. | ||
Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol. 2010;52(2):252–257. | ||
Stolzenberg-Solomon RZ, Qiao Y-L, Abnet CC, et al. Esophageal and gastric cardia cancer risk and folate-and vitamin B12-related polymorphisms in Linxian, China. Cancer Epidemiol Biomarkers Prev. 2003;12(11):1222–1226. | ||
Jarmalaite S, Andrekute R, Scesnaite A, Suziedelis K, Husgafvel-Pursiainen K, Jankevicius F. Promoter hypermethylation in tumour suppressor genes and response to interleukin-2 treatment in bladder cancer: a pilot study. J Cancer Res Clin Oncol. 2010;136(6):847–854. | ||
Okamoto T, Tsuburaya A, Yanoma S, et al. Inhibition of peritoneal metastasis in an animal gastric cancer model by interferon-gamma and interleukin-2. Anticancer Res. 2002;23(1A):149–153. | ||
Terao S, Shirakawa T, Goda K, Kamidono S, Fujisawa M, Gotoh A. Recombinant interleukin-2 enhanced the antitumor effect of ADV/RSV-HSV-tk/ACV therapy in a murine bladder cancer model. Anticancer Res. 2005;25(4):2757–2760. | ||
Matesanz F, Delgado C, Fresno M, Alcina A. Allelic selection of human IL-2 gene. Eur J Immunol. 2000;30(12):3516–3521. | ||
Williams TM, Eisenberg L, Burlein JE, et al. Two regions within the human IL-2 gene promoter are important for inducible IL-2 expression. J Immunol. 1988;141(2):662–666. | ||
John S, Turner D, Donn R, et al. Two novel biallelic polymorphisms in the IL-2 gene. Eur J Immunogenet. 1998;25(6):419–420. | ||
Hu XB, Ouyang LZ, Tang LL. Interleukin-2 gene polymorphisms and prognosis of breast cancer. Genet Test Mol Biomarkers. 2013;17(6):453–457. | ||
Song N, Han S-H, Lee K-M, et al. Genetic variants in interleukin-2 and risk of lymphoma among children in Korea. Asian Pac J Cancer Prev. 2012;13(2):621–623. | ||
Berković MC, Jokic M, Marout J, Radosevic S, Zjacic-Rotkvic V, Kapitanovic S. IL-2 −330 T/G SNP and serum values-potential new tumor markers in neuroendocrine tumors of the gastrointestinal tract and pancreas (GEP-NETs). J Mol Med. 2010;88(4):423–429. | ||
Savage SA, Abnet CC, Haque K, et al. Polymorphisms in interleukin-2, -6, and -10 are not associated with gastric cardia or esophageal cancer in a high-risk chinese population. Cancer Epidemiol Biomarkers Prev. 2004;13(9):1547–1549. | ||
Peng Q, Li H, Lao X, et al. Association of IL-2 polymorphisms and IL-2 serum levels with susceptibility to HBV-related hepatocellular carcinoma in a Chinese Zhuang population. Infect Genet Evol. 2014;27:375–381. | ||
Bei C-H, Bai H, Yu H-P, et al. Combined effects of six cytokine gene polymorphisms and SNP-SNP interactions on hepatocellular carcinoma risk in Southern Guangxi, China. Asian Pac J Cancer Prev. 2014;15(16):6961–6967. | ||
Zhao H, Wang R. IL-2 −330T/G polymorphism and cancer risk: a meta-analysis. Onco Targets Ther. 2015;8:1753–1760. | ||
Howell WM, Turner SJ, Theaker JM, Bateman AC. Cytokine gene single nucleotide polymorphisms and susceptibility to and prognosis in cutaneous malignant melanoma. Eur J Immunogenet. 2003;30(6):409–414. | ||
Wells G, Shea B, O’connell D, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Ottawa:Ottawa Hospital Research Institute; 2000 [cited October 19, 2009]. Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.htm. Accessed July 1, 2015. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies. J Natl Cancer Inst. 1959;22(4):719–748. | ||
Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10(1):101–129. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.











