Back to Journals » Infection and Drug Resistance » Volume 12
Association between cell growth and vancomycin resistance in clinical community-associated methicillin-resistant Staphylococcus aureus
Authors Yamaguchi T , Ando R, Matsumoto T , Ishii Y, Tateda K
Received 21 March 2019
Accepted for publication 21 June 2019
Published 1 August 2019 Volume 2019:12 Pages 2379—2390
DOI https://doi.org/10.2147/IDR.S209591
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Tetsuo Yamaguchi,1,2 Rina Ando,2 Tetsuya Matsumoto,2 Yoshikazu Ishii,1 Kazuhiro Tateda1
1Department of Microbiology and Infectious Diseases, Toho University School of Medicine, Tokyo, Japan; 2Department of Microbiology, Tokyo Medical University, Tokyo, Japan
Objectives: We investigated the association between the rate of cell growth and vancomycin (VAN) resistance by comparing the genomic and phenotypic characteristics of different sized colonies isolated from a single origin.
Methods: Vancomycin-intermediate Staphylococcus aureus (VISA) strain TMUS2136 was isolated from the pus of a patient with a brain abscess and a chronic subdural abscess. This strain grew slowly and formed colonies poorly on agar plates within 24 h of incubation. However, variously sized colonies were observed after 48 h of incubation. We isolated five strains from different sized colonies and compared their VAN susceptibility and genomic sequences using next-generation sequencing.
Results: The five strains showed different rates of growth and susceptibilities to VAN (range of MIC after 48 h of incubation; 3–5 mg/L), and slower growing strains tended to be more VAN resistant. Deletion of the spdC gene and SNPs in the sarA gene was confirmed in all five strains and six SNPs in other genes (including mreC, hssS, prs, SA2339, sun, and SA1132) were confirmed when comparing the five strains.
Conclusions: Deletion of the spdC gene, a novel virulence factor of S. aureus, and SNPs in the sarA gene may be associated with VAN resistance. It was hypothesized that any of the six genes in which SNPs were detected could affect cell growth rate and VAN resistance in slow-VISA strains.
Keywords: next generation sequencing, MRSA, VISA, vancomycin
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) was first isolated in 1960 in England, just after methicillin had become commercially available. MRSA is a derivative of S. aureus that gained resistance to β-lactam antibiotics by acquiring the mecA gene (or mecC gene) in its chromosomal DNA.1 It has become endemic in most hospitals and healthcare facilities in industrialized countries since the 1980s (termed healthcare-associated MRSA, HA-MRSA).2,3 Furthermore, community-associated MRSA (CA-MRSA) infections occurring in otherwise healthy individuals, including children, appeared in the 1990s,4,5 suggesting that these bacterial strains have greater virulence than traditional HA-MRSA strains. Since the mid-2000s, MRSA has also been associated with livestock exposure (livestock-associated MRSA, LA-MRSA),6 and has thus become a health threat in a variety of environments.
In addition to virulence factors, a key factor that makes it more difficult to cure MRSA infection is the diversity of mechanisms of antimicrobial resistance. The year 2020 marks the 60th anniversary of our continuous battle against MRSA. MRSA has acquired resistance to practically all the antibiotics brought into clinical use in the past 50 years, including vancomycin (VAN).7,8 According to the minimum inhibitory concentrations (MICs) of VAN, S. aureus strains are classified into vancomycin susceptible S. aureus (VSSA) (minimum inhibitory concentration [MIC] ≤2 mg/L), vancomycin intermediate S. aureus (VISA) (MIC 4–8 mg/L), and vancomycin resistant S. aureus (VRSA) (MIC ≥16 mg/L). VRSA is a highly VAN-resistant strain that acquired the vanA gene through conjugated transfer, although fortunately it has not been detected in Japan.9 It is known that the mechanism of resistance of VISA differs from that of VRSA, and VISA strains are often detected around the world, including Japan.10 VISA acquires VAN resistance by thickening of the cell wall. It was reported that one or several single nucleotide polymorphisms (SNPs) in cell wall formulation genes, for example SNPs in walK, walS, or walR, cause thickening of the cell wall and result in reduced sensitivity to VAN.10 However, it is sometimes difficult to detect clinically relevant VISA strains with precision, because there are cases where the MIC of the clinical VISA strain is under 4 mg/L, including hetero-VISA and slow growing VISA (slow-VISA).11,12,13 The infectious disease caused by VISA, including hetero-VISA and slow-VISA, may show VAN-treatment resistance, and therefore it is important to detect VISA strains reliably in clinical settings.
We have obtained the VISA strain isolated from a subdural abscess.14 This clinical isolate grows slowly, thus the MICs of VAN after 24 and 48 h of incubation were 2 mg/L (sensitive) and 4 mg/L (intermediate), respectively. According to the Clinical & Laboratory Standards Institute (CLSI) criteria (24 h incubation),15 this isolate was therefore judged as VSSA, not VISA. However, the present case showed resistance to VAN treatment in a clinical setting. It was difficult to judge the resistance of this organism because of its slow growth; however, we conducted population analysis of this isolate and determined that it was a VISA strain. Additionally, this isolate grows slowly and forms colonies poorly on agar plates within 24 h of incubation, although it produces various sized colonies after 48 h of incubation. Several strains obtained from a variety of different sized colonies showed different VAN sensitivities and growth rates when compared with each other. It was thought that these strains were the same clone having the same origin; however, it is a problem clinically if VAN sensitivity changes depending on which colony is picked.
In this study, we compared the genomic and phenotypic characteristics of these strains obtained from a variety of different sized colonies sharing the same origin. The purpose of this study was to clarify the association between cell growth rate (colony size), VAN resistance, and genome sequence.
Materials and methods
Bacterial strains
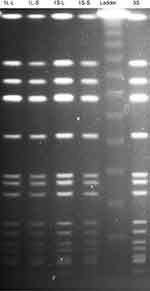
Vancomycin-intermediate Staphylococcus aureus (VISA) strain TMUS2136 was isolated from the pus of a 77-year-old Japanese patient with brain and chronic subdural abscesses.14 He had multiple administrations of anti-MRSA antimicrobial agents, including VAN (3 weeks) and linezolid (LZD) (2 weeks), for a MRSA subdural abscess 2 years previously, followed by minocycline (MINO) for a chronic subdural abscess for 2 years (Figure 1). This strain grew slowly and formed colonies poorly on agar plates within 24 h of incubation, although colonies of various sizes were observed after 48 h incubation. We purified five different sized colonies (designated 1L-L, 1L-S, 1S-L, 1S-S, and 3S in descending order of size) of strain TMUS2136 on agar plates after repeating incubation twice (Figure 2). We subsequently analyzed these five strains and four additional control MRSA strains (USA300 clone BAA1556, New York/Japan clone N315, hetero-VISA strain Mu3, and VISA strain Mu50). Pathogen protocols were approved by Toho University Safety Committee for Pathogens (approval number 17-55-58).
Growth rate of strains
The growth rate of these strains was determined by their absorbance values. Each strain was cultured on BHI agar overnight. Then 0.5 McFarland suspensions from colonies on the BHI agar were diluted 200 times into 6 mL of fresh BHI broth in an L-shaped tube and were incubated at 35 °C with mixing by inversion at 25 rpm in an automatic photorecording incubator (TVS062CA, Advantec, Osaka, Japan). The optical density (OD) at 660 nm was automatically monitored and recorded every 10 min. For doubling-time determinations, the OD versus time was plotted for each strain in the exponential growth phase. The doubling times were then calculated as follows: [(t2 - t1) x log 2]/(log OD660 at t2 ˗ log OD660 at t1).16 Doubling time was measured in at least three independent experiments and was used for statistical analysis. One-way ANOVA followed by Tukey’s multiple comparison test were performed using GraphPad Prism (version 8, GraphPad Software Inc., San Diego, CA, USA). P-values <0.0001 were considered statistically significant.
VAN susceptibility tests
The VAN MIC of strains at several time points was determined by an Etest using Etest strips (bioMerieux, Marcy-l’Etoile, France) and MHA plates (Eiken Chemical, Tokyo, Japan). The time points were 24, 48, 72, and 96 h after incubation.
Population analysis
Analysis of resistant subpopulations of bacteria (population analysis) was performed by spreading 50 μL of the starting cell suspension (prepared by diluting overnight cultures to an OD of 0.1, as determined by a MicroScan Turbidity Meter (SIEMENS, Munich, Germany)) and its serial diluents over BHI agar plates containing various concentrations of VAN. The plates were incubated at 35 °C for 72 h, and the number of mature colonies was counted at 24, 48, and 72 h. The number of resistant cells contained in the starting cell suspension was calculated and plotted on a semi-logarithmic graph.
Pulsed-field gel electrophoresis (PFGE)
All strains were examined by pulsed-field gel electrophoresis. After digestion of the genomic DNA with SmaI (New England Biolabs, Ipswich, MA, USA), the restriction fragments were separated using a temperature-controlled CHEF DR III system (Bio-Rad, Hercules, CA, USA) at 6.0 V for 24 h at switch times ramped from 5 to 60 s. After staining with ethidium bromide, the fragments were visualized using an UV transilluminator.
Whole-genome sequencing and identification of mutations
Each of the strains were cultured overnight in heart infusion broth (Eiken Chemical Co.) and the cells were harvested by centrifugation at 8,000× g for 3 min. Bacterial genomic DNA was then extracted using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) with lysostaphin (Wako, Osaka, Japan). Genomic DNAs were used as a template for whole-genome sequencing by next-generation sequencing (NGS) using MiSeq (Illumina, San Diego, CA, USA). DNA-libraries for NGS were prepared with Nextera XT DNA sample preparation kits (Illumina), in accordance with the manufacturer’s protocol. Pools of five samples were sequenced on the Illumina MiSeq platform, and 250-bp end reads were generated. Reads from bacterial strains were assembled using CLC Genomics Workbench v 11.0.1 software (Qiagen), and obtained contigs were analyzed using the SCCmecFinder service (https://cge.cbs.dtu.dk/services/SCCmecFinder/)17 and the Multi-Locus Sequence Typing (MLST) service (https://cge.cbs.dtu.dk/services/MLST/) on the Center for Genomic Epidemiology homepage. Furthermore, reads from bacterial strains were mapped to the whole-genome reference sequence of N315 strain (accession no. NC_002745.2) and BAA1556 strain (accession no. NC_007793.1, the same sequence type strain as TMUS2136; ST8) using CLC Genomics Workbench v 11.0.1 software (Qiagen). The mutations were identified by comparing the draft genome sequences from isolated strains against the reference sequences. The sequence data for isolates were deposited in the DNA Data Bank of Japan (DDBJ) under accession numbers SAMD00115769 (3S), SAMD00115770 (1S-S), SAMD00115771 (1S-L), SAMD00115772 (1L-S), and SAMD00115773 (1L-L), respectively.
Results
Genome type of isolates
All five strains (1L-L, 1L-S, 1S-L, 1S-S, and 3S) showed the same banding pattern on PFGE (Figure 3). The genome types of these strains were ST8, SCCmec type IV, spa type t008, agr type I, PVL gene negative, and TSST-1 gene positive, the same genome type as the CA-MRSA/J clone that is the dominant clone in Japan.18 Notably, this clone is isolated from not only community-associated settings, but also hospital-associated settings.19,20
Growth rate of strains
The growth rate curves of the isolates are shown in Figure 4. Strain 3S obtained from the smallest colony grew the slowest, and strain 1L-L obtained from the largest colony grew the fastest. The slower the growth rate, the smaller the colony. The mean doubling times of BAA1556, N315, Mu3, Mu50, 1L-L, 1L-S, 1S-L, 1S-S, and 3S were 40.7, 45.8, 52.5, 57.0, 62.8, 66.8, 69.6, 115.1, and 124.0 min, respectively. Compared with BAA1556, N315, Mu3, Mu50, 1L-L, 1L-S, and 1S-L, 1S-S and 3S showed significantly longer doubling times (P<0.0001). 1S-S and 3S required 2 days incubation for MIC measurement by the Etest, and the mean doubling time was over 100 min. Strains with a doubling time of more than 100 min may be considered slow growing.
VAN susceptibility tests
The VAN MICs of strains used in this study are shown in Table 1. The MIC of strain 1L-L was 3 mg/L after 24 h incubation, and this did not change with additional incubation time (until after 96 h). The MIC of strain 1L-S was 2 mg/L after 24 h incubation; however, it changed to 3 mg/L after over 48 h incubation. The MICs of strains 1S-L, 1S-S, and 3S were difficult to determine because the growth of these strains could not be observed after 24 h incubation. However, after over 48 h incubation, the MIC of strains 1S-L and 1S-S was 4 mg/L. The MIC of strain 3S was 3 mg/L after 48 h incubation, increasing to 4 mg/L after 72 h incubation, and up to 5 mg/L after 96 h incubation. Interestingly, the slower the growth rate, the higher the MIC. The MIC of strains Mu3 and Mu50 were that of typical hetero-VISA and VISA strains, respectively, and did not change after over 48 h incubation (Table 1). Strain 3S required 96 h growth before the VAN MIC could be determined, thus it was hypothesized that this strain should be categorized as a slow-VISA.
 |
Table 1 VAN minimum inhibitory concentration (MIC) of isolates, as determined by an Etest (mg/L). |
Population analysis
The results of population analysis are shown in Figure 5. It is known that VISA strains, including strains Mu3 and Mu50, grow slowly. Therefore, colony counts for population analysis were conducted after 48 h incubation. The growth of all five strains was confirmed on BHI agar plates with 4 mg/L VAN after 48 h incubation. The definition of hetero-VISA was satisfied, but these five strains were not VISA because the MICs were <4 mg/L. The population of strain 1L-L on agar plates containing various VAN concentrations did not change after 24, 48, and 72 h. The population of strains 1S-L, 1S-S, and 3S after 24 h incubation could not be counted because colonies of these strains were not observed after only 24 h incubation. The population of strains 1S-S and 3S were thus counted after 48 h, and changes were observed between 24, 48, and 72 h incubation. Notably, the population of strain 3S changed significantly, thus it is important for population analysis to be undertaken after over 72 h incubation.
Genes implicated in the VAN intermediate phenotype in strain TMUS2136
The major genes implicated in the VAN intermediate phenotype in previous reports are listed in Table 2.21–25 Compared with NC_007793 (BAA1556/ST8/VSSA), amino acid changes were detected in agrC, sarA, and trfB; however, the sequences in agrC and trfB were the same as NC_002745 (N315/ST5/VSSA). These data suggest that the SNPs in agrC and trfB were not associated with VAN resistance. Additionally, we confirmed the sequence of the spdC gene that is a novel virulence factor of S. aureus.26 Compared with NC_002745, deletion of the spdC gene in TUM2136 was confirmed. Expression of spdC is positively regulated by the walKR system and spdC negatively controls walKR regulon genes, thus deletion of spdC may be associated with VAN resistance.
 |
Table 2 Genes associated with the VISA phenotype and nucleotide changes in genes of strain TMUS2136 compared with BAA1556 (NC_007793) |
Mutations according to comparisons with the whole genome sequences mapping to reference strains
Comparing the whole genome sequences of the five strains obtained from strain TMUS2136 detected SNPs in only six genes (Table 3). SNPs in mreC and hssS were detected only in the strain 1L-L genome, while SNPs in prs were detected in the genomes of strains 1L-L and 1L-S. SNPs in porB were detected only in the 3S genome, while SNPs in sun were detected in the genomes of strains 1L-S, 1S-L, 1S-S, and 3S. SNPs in SA2339 were detected only in the 1S-L genome. Notably, strains 1L-L and 1L-S both had SNPs in prs, compared with the slower growing strains including 1S-L, 1S-S, and 3S. Thus, prs may affect the growth rate, cell wall formulation, and VAN resistance of MRSA. The extremely slow growing strain 3S had SNPs in porB, which may have relevance to slow-VISA.
 |
Table 3 Mutations according to comparisons with whole genome sequences mapping to N315 (NC_002745) |
Discussion
In this study, we demonstrated that strains with slower growth and smaller colony formation tended to be resistant to VAN. Several genes, such as spdC and prs, were detected as new genes implicated in VAN resistance and cell growth. Slow-VISA strains are extremely slow growing and need longer incubation times to observe colony formation. Therefore, it is difficult to isolate such strains and to analyze accurate antimicrobial susceptibility by standard methods according to the guidelines of the CLSI M7.27
Initially, an amino acid change in the sarA gene and a deletion of the spdC gene in VISA strain TUM2136 were confirmed by comparison with a reference strain. The sarA gene encodes an important protein, SarA, which positively regulates the agr gene and is associated with the expression of surface proteins, capsular polysaccharides, and toxins.28 The sarA gene has been previously implicated as a VAN resistance gene.29 It is highly likely that SNPs in sarA influence VAN resistance. Moreover, SpdC is a novel virulence factor reported in recent years, and negatively controls WalKR regulon genes.26 Additionally, it is known that the WalKR system is involved in controlling cell wall metabolism and is associated with VAN resistance. Therefore, it was hypothesized that the deletion of spdC affected the dynamics of WalKR and lead to VAN resistance.
Furthermore, a total of six SNPs were detected by comparing the genomes of the isolated strains and these may be associated with MRSA growth and resistance to VAN. The transmembrane protein MreC encoded by the mreC gene is thought to be required to maintain the shape of rod-shaped bacteria.30,31 However, it has been reported that mreC is not essential in MRSA and does not affect cell morphology.32 Similarly, HssS is part of the heme sensing system (HssRS) that controls the heme regulated transporter efflux pump (HrtAB), and it is unclear how it is associated with synthesis of the cell wall and VAN resistance.33 The prs gene encodes ribose-phosphate pyrophosphokinase and it has been reported that a mutation in this gene causes daptomycin resistance.34 The mechanism by which SA2339, sun, and SA1132 operate remains unclear because there are no reports regarding their association with VAN resistance or cell growth. Among the six genes, it was suggested that prs was most likely responsible for the cell growth rate and VAN susceptibility because SNPs in the prs gene were detected during comparison between strains 1L-S and 1S-L, which represent the borderline of VAN susceptibility. Furthermore, the SA1132 gene may be responsible for slow-VISA because SNPs in the SA1132 gene were confirmed only in strain 3S. Notably, an association between this gene and VAN resistance has not been reported previously.
Importantly, questions remain regarding why various sized colonies were purified from a single clone isolated from a single patient. This growth phenomenon regarding S. aureus has been reported upon previously, including the description of hetero-VISA and small colony variants; however, the mechanisms responsible were not clear. The “heterogeneity” of VISA is explained by population analysis, in which over 100 colonies were observed on 4 mg/L VAN agar plates inoculated with 108 bacteria after 48 h incubation.10 This indicates that there are at least two types of strains present in the bacterial suspension from a single colony of an isolate, one of which is susceptible and the other is resistant. Critically, a single colony from a purified culture is generally believed to be formed by a pure single strain. This concept is widely accepted in microbiology; however, it does not support the idea of heterogeneity in VISA strains, with a single colony originally containing several types of strains. A concept that may explain the fact that a single colony from a strain includes several subtypes of strains is genetic mutation. If mutants emerge from a strain during the process of colony formation, the environment-compatible mutants may become predominant in that colony. Our hypothesis is that the TMUS2136 strain acquired VAN intermediate resistance because it was exposed to VAN in the host body. The mechanism of resistance was thickening of the cell wall that resulted in slower cell division and cell growth. We think that this is the case for 3S strain. By analysis of colony forming units or bacterial suspensions, it was difficult to clarify whether the mechanism was colony heterogeneity or de novo mutation. However, SNPs in six genes were detected between the five strains in this study, and mutants could emerge either in the host body or during the process of purified incubation. To clarify the specific mechanisms, we need new methods that can observe a single bacterial cell and analyze each cell separately.
In clinical settings, there may be some cases where strains with various growth rates and/or antibiotic susceptibilities are isolated from a single source like the present case. In these cases, various sized colonies are observed and the smallest colonies are difficult to detect because larger colonies dominant as a result of their fast growth, which may mask the smaller colonies. Notably, a strain that requires over 24 h incubation for observation of colonies such as strain 3S cannot be detected under normal incubation conditions. This suggests that slower and more antibiotic-resistant strains are difficult to detect, thus it is necessary to consider longer incubations to detect the smaller colonies.
TMUS2136 strain is a ST8/SCCmec type IV/TSST-1 positive VISA strain that is the dominant Japanese CA-MRSA clone, also called CA-MRSA/J.18 This type of clone has been isolated in not only community-associated settings, but also hospital-associated settings in Japan.19,20 Notably, many such clones were isolated from bacteremia cases and were thought to be highly virulent strains. CA slow-VISA presents such a threat because of its high virulence, resistance to VAN, and difficulty of detection. Thus, we need to focus on preventing the emergence and spread of slow-VISA, especially CA slow-VISA.
There are several limitations to the present study. First, TMUS2136 strain was isolated from a patient two years after receiving initial VAN treatment, and MRSA isolates from the initial stage of the subdural abscess were not purified and stored. Therefore, its growth characteristics were not clarified and it is unclear when and why slow-VISA emerged. Notably, it is impossible to know what factors induced VAN resistance and resulted in the emergence of slow-VISA as various antimicrobial agents, including VAN, LZD, and MINO, were administered. Second, we did not directly prove the relationship between the genes detected in this study and VAN resistance using a gene knock-out strain. It was difficult to clarify whether the mechanism of diversity in a single colony was a result of colony heterogeneity or de novo mutation, because purification and analysis of a single bacterial cell was not possible in our laboratory. If such technology were available, further investigations into the relationship between colony heterogeneity, VAN resistance, and gene mutations would be conducted.
In conclusion, we have demonstrated that SNPs in sarA, prs, or SA1132 and/or deletion of spdC may be implicated as a cause of VAN resistance and the slow growth of MRSA. Slower growing strains tend to be VAN resistant, so the smallest colonies (after a long incubation period) should be tested for antimicrobial susceptibility to detect VAN-resistant organisms. Importantly, overlooking VAN-resistant CA-MRSA may lead to severe infections and resistance to treatment. Therefore, it is necessary to establish a new method to detect slow-VISA and a new therapeutic strategy to combat it. Further investigations should be performed to clarify the mechanisms regulating cell growth and VAN resistance in MRSA.
Ethics approval
Written informed consent was obtained from the patient’s family for publication of this study.
Acknowledgments
We thank Miki Matsuo, Yuki Katayama, and Keiichi Hiramatsu for their total support of this report. We thank Simon Teteris, PhD, from the Edanz Group, for editing the English text of a draft of this manuscript. This work was supported by a GlaxoSmithKline Japan Research Grant and also by grants provided by Takeda Science Foundation.
Disclosure
Current affiliation for Tetsuya Matsumoto is at Department of Infectious Diseases, International University of Health and Welfare, Chiba, Japan. The authors report no conflicts of interest in this work.
References
1. Jevons MP. “Celbemom”-resistant staphylococci. Br Med J. 1961;1:124–125.
2. Hiramatsu K. Molecular evolution of MRSA. Microbiol Immunol. 1995;39:531–543.
3. Enright MC, Robinson DA, Randle G, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A. 2002;99:7687–7692. doi:10.1073/pnas.122108599
4. CDC. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus–minnesota and North Dakota, 1997-1999. JAMA. 1999;282:1123–1125.
5. Deleo FR, Otto M, Kreiswirth BN, et al. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi:10.1016/S0140-6736(09)61999-1
6. Ferber D. Infectious disease. From pigs to people: the emergence of a new superbug. Science. 2010;329:1010–1011. doi:10.1126/science.329.5995.1010
7. Hiramatsu K, Hanaki H, Ino T, et al. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi:10.1093/jac/40.1.135
8. Chang S, Sievert DM, Hageman JC, et al. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. 2003;348:1342–1347. doi:10.1056/NEJMoa025025
9. Sievert DM, Rudrik JT, Patel JB, et al. Vancomycin-resistant Staphylococcus aureus in the United States, 2002-2006. Clin Infect Dis. 2008;46:668–674.
10. Howden BP, Davies JK, Johnson PD, et al. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. 2010;23:99–139. doi:10.1128/CMR.00042-09
11. Howden BP, Peleg AY, Stinear TP. The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect Genet Evol. 2014;21:575–582. doi:10.1016/j.meegid.2013.03.047
12. Katayama Y, Azechi T, Miyazaki M, et al. Prevalence of slow-growth vancomycin nonsusceptibility in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2017;61:
13. Matsuo M, Hishinuma T, Katayama Y, Hiramatsu K. A mutation of RNA polymerase β’ subunit (RpoC) converts heterogeneously vancomycin-intermediate Staphylococcus aureus (hVISA) into “slow VISA”. Antimicrob Agents Chemother. 2015;59:4215–4225. doi:10.1128/AAC.00135-15
14. Kino H, Suzuki H, Yamaguchi T, et al. Central nervous system infection caused by vancomycin-intermediate Staphylococcus aureus (SCCmec type IV, ST8). J Infect Chemother. 2014;20:643–646. doi:10.1016/j.jiac.2014.06.008
15. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; 27th Informational Supplement. Wayne, PA: CLSI document M100-S27 Clinical and Laboratory Standards Institute; 2017.
16. Matsuo M, Hishinuma T, Katayama Y, et al. Mutation of RNA polymerase beta subunit (rpoB) promotes hVISA-to-VISA phenotypic conversion of strain Mu3. Antimicrob Agents Chemother. 2011;55:4188–4195. doi:10.1128/AAC.00398-11
17. Kaya H, Hasman H, Larsen J, et al. sccmecfinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere. 2018:3:
18. Iwao Y, Ishii R, Tomita Y, et al. The emerging ST8 methicillin-resistant Staphylococcus aureus clone in the community in Japan: associated infections, genetic diversity, and comparative genomics. J Infect Chemother. 2012;18:228–240. doi:10.1007/s10156-012-0379-6
19. Yamaguchi T, Okamura S, Miura Y, et al. molecular characterization of community-associated methicillin-resistant Staphylococcus aureus isolated from skin and pus samples of outpatients in Japan. Microb Drug Resist. 2015;21:441–447. doi:10.1089/mdr.2014.0153
20. Miura Y, Yamaguchi T, Nakamura I, et al. Epidemiological trends observed from molecular characterization of methicillin-resistant Staphylococcus aureus isolates from blood cultures at a Japanese university hospital, 2012-2015. Microb Drug Resist. 2018;24:70–75. doi:10.1089/mdr.2017.0008
21. McGuinness WA, Malachowa N, DeLeo FR. Vancomycin resistance in Staphylococcus aureus. Yale J Biol Med. 2017;90:269–281.
22. Rishishwar L, Petit RA
23. Herbert S, Bera A, Nerz C, et al. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 2007;3:e102. doi:10.1371/journal.ppat.0030102
24. Hafer C, Lin Y, Kornblum J, et al. Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:5845–5851. doi:10.1128/AAC.01139-12
25. Matsuo M, Cui L, Kim J, et al. Comprehensive identification of mutations responsible for hVISA-to-VISA conversion in laboratory-generated VISA strains from hVISA clinical strain Mu3. Antimicrob Agents Chemother. 2013. doi:10.1128/AAC.00425-13.
26. Poupel O, Proux C, Jagla B, et al. SpdC, a novel virulence factor, controls histidine kinase activity in Staphylococcus aureus. PLoS Pathog. 2018;14:e1006917. doi:10.1371/journal.ppat.1006917
27. CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically.
28. Reyes D, Andrey DO, Monod A, et al. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J Bacteriol. 2011;193:6020–6031. doi:10.1128/JB.05436-11
29. Trotonda MP, Xiong YQ, Memmi G, et al. Role of mgrA and sarA in methicillin-resistant Staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J Infect Dis. 2009;199:209–218. doi:10.1086/595740
30. Carballido-Lopez R, Formstone A. Shape determination in Bacillus subtilis. Curr Opin Microbiol. 2007;10:611–616. doi:10.1016/j.mib.2007.09.008
31. Divakaruni AV, Loo RR, Xie Y, et al. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc Natl Acad Sci U S A. 2005;102:18602–18607. doi:10.1073/pnas.0507937102
32. Tavares AC, Fernandes PB, Carballido-Lopez R, et al. MreC and MreD proteins are not required for growth of Staphylococcus aureus. PLoS One. 2015;10:e0140523. doi:10.1371/journal.pone.0140523
33. Stauff DL, Torres VJ, Skaar EP. Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J Biol Chem. 2007;282:26111–26121. doi:10.1074/jbc.M703797200
34. Song Y, Rubio A, Jayaswal RK, et al. Additional routes to Staphylococcus aureus daptomycin resistance as revealed by comparative genome sequencing, transcriptional profiling, and phenotypic studies. PLoS One. 2013;8:e58469. doi:10.1371/journal.pone.0058469
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.