Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 15
Association Among MIF, IFIH1, and IL6 Gene Polymorphisms and Non-Segmental Vitiligo in a Chinese Han Population
Authors Wang D, Min S, Lin X, Jiang G
Received 6 April 2022
Accepted for publication 13 July 2022
Published 10 August 2022 Volume 2022:15 Pages 1597—1609
DOI https://doi.org/10.2147/CCID.S369418
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
Danfeng Wang,1,2 Shuhui Min,1,2 Xiao Lin,1,2 Guan Jiang1,2
1Department of Dermatology, Affiliated Hospital of Xuzhou Medical University, Xuzhou, People’s Republic of China; 2First Clinical Medical College, Xuzhou Medical University, Xuzhou, People’s Republic of China
Correspondence: Guan Jiang, Department of Dermatology, Affiliated Hospital of Xuzhou Medical University; First Clinical Medical College, Xuzhou Medical University, Xuzhou, People’s Republic of China, Email [email protected]
Objective: The aim of the present study was to investigate the association of single-nucleotide polymorphisms (SNPs) in the macrophage migration inhibiting factor (MIF), interferon-induced Helicase C domain 1 (IFIH1), interleukin-6 (IL6) genes, circulating levels with non-segmental vitiligo (NSV) susceptibility in the Chinese population, and to analyze the relationships between gene polymorphisms and clinical characteristics of vitiligo.
Methods: In this study, genotyping was conducted in 155 patients with NSV and 117 unaffected controls using polymerase chain reaction and snapshot technique. Serum concentrations were determined by ELISA kit.
Results: There were strong associations between IFIH1 H843R and IL6-572G/C polymorphisms and NSV susceptibility (p = 0.013; p = 0.009). In contrast to previous studies, we found no significant difference in the MIF-173G/C polymorphism between the two groups. In addition, the frequency of allelic distribution for MIF-173G/C in patients with active NSV was significantly higher than stable NSV (p = 0.011), and IFIH1 H843R with early-onset (≤ 20), active or family history of NSV was significantly higher than late-onset (> 20), stable or no family history of NSV (p = 0.033; p = 0.045; p = 0.039). Serum concentrations of MIF were higher in patients with active NSV, serum IFIH1 and IL6 concentrations were related to the presence of polymorphisms in patients with NSV (p = 0.009; p = 0.011).
Conclusion: Our results suggested that IFIH1 H843R and IL6-572G/C gene polymorphisms and expression levels are obviously correlated with the onset of NSV. MIF-173G/C allele and serum concentrations may be associated with active NSV, and IFIH1 H843R allele may be associated with youth, active or family history of NSV.
Keywords: vitiligo, MIF, IFIH1, IL6, single nucleotide polymorphism
Introduction
Vitiligo is the most common acquired chronic depigmentation disease that is characterized by selective destruction of functional melanocytes in the skin or hair follicles. Vitiligo affects 0.5% to 2% of the population worldwide,1 and the incidence of vitiligo in China is 0.5%.2 Although the disease is not life-threatening, it can lead to disfigurement and seriously affect the patient’s appearance. In addition, vitiligo is associated with a variety of autoimmune diseases,3 which can have a serious impact on mental health and quality of life. Vitiligo is a complex disease that combines genetic and environmental factors as well as metabolic and altered inflammatory and immune responses.4 Currently, although many vitiligo-related pathogenic genes have been identified and preliminarily confirmed to be related to vitiligo, the exact genetic mechanism of vitiligo cannot be fully explained.
Single-nucleotide polymorphisms (SNPs) of genes refer to the substitution of a single nucleotide at a specific position in the genome and account for most human heritable variations. There is convincing evidence that proinflammatory cytokines play a vital role in the initiation of vitiligo lesions.5 Several SNPs of inflammatory cytokines have been demonstrated to be associated with vitiligo, but contradictory results exist among different ethnic groups.
MIF was initially described as a chemotactic lymphocytokine and a negative regulator of the immunosuppressive actions of glucocorticoids. Subsequently, MIF has been described as a proinflammatory factor with upstream regulatory roles in innate and adaptive immunity.6 It has been shown to play a crucial role in several types of immune and autoimmune diseases.7,8 MIF is mainly derived from macrophages and T cells and is secreted in response to several stimuli, including lipopolysaccharide (LPS), tumor necrosis factor (TNF)‐α, hypoxia, and oxidative stress.9–11 In the presence of inflammation, MIF allows for prolonged inflammation through the release of other pro-inflammatory cytokines.12 MIF-173G/C(rs755622 [RefSeq NG_012099.1]) is located in the promoter region of chromosome 22. One study on the western Mexico population demonstrated that MIF-173G/C will become an extremely attractive target for vitiligo treatment.13 However, a similar conclusion has not been confirmed in vitiligo patients of different populations.
IFIH1, also known as melanoma differentiation-associated protein 5 (MDA5), is a cytosolic innate immune receptor that recognizes viral RNA and activates IFN regulatory factor 3 (IRF3) and the proinflammatory transcription factor nuclear factor-kB (NF-kB), to induce inflammation and other antiviral genes.14 During inflammatory conditions and infections, viral RNA or RNA mimics and lipopolysaccharide (LPS) can activate IFIH1 production by stimulating distinct signaling pathways.15 IFIH1 is an early type I interferon (IFN) β response gene, which plays a critical role in initiating antiviral innate immunity and modulating subsequent adaptive immunity.16 Several genome-wide association studies (GWAS) have confirmed that IFIH1 is a vitiligo susceptibility gene.17,18 A recent GWAS showed that a single nucleotide polymorphism IFIH1 H843R(rs3747517 [RefSeq NG_011495.1]) in the exon of chromosome 2 was associated with vitiligo in a Chinese population.19 In order to further study the etiology of vitiligo, we conducted the genotype of phenotype of this locus in depth.
IL6 is a pleiotropic cytokine, produced by macrophages, T cells and B cells, was initially designated as a B cell stimulator that promoted maturity of B-cells and the expression of immunoglobulins.20 During inflammatory conditions and infections, certain bacterial LPSs, interleukin-1b (IL-1b) and tumor necrosis factor (TNF), are important stimuli for IL6 production.21 Through different signaling pathways, IL6 induces acute phase protein generation, contributing to the development of multiple inflammatory and autoimmune diseases.22 As a common clinical inflammatory factor, IL6 has been shown to be significantly elevated in the body fluids of patients with vitiligo.23 However, SNPs in the IL6 gene have been identified in various diseases, but few studies concerning IL6 gene promoter polymorphism at position −572G/C(rs1800796 [RefSeq NG_011640.1]) were conducted on patients with vitiligo.
Therefore, it is of significance to explore the pathogenesis of vitiligo, especially the immunological and genetic backgrounds, for the development of new therapeutic targets for vitiligo. The current study aimed to measure the serum MIF, IFIH1 and IL6 level and the role of the SNP (rs755622, rs3747517 and rs1800796) as a risk for the susceptibility to vitiligo, and to determine its correlation with disease activity and severity of vitiligo.
Materials and Methods
Patients
A total of 155 patients with NSV and 117 sex- and age-matched unaffected controls from the same geographical area were enrolled in the present study. According to the revised classification of vitiligo by the Vitiligo Global Issues Consensus Conference (VGICC) published in 2012,24 there are four recognized clinical forms of vitiligo: non-segmental, segmental, mixed, and unclassified. We selected the most common type of NSV for the study, and NSV including acrofacial, generalized, mucosal, and universal vitiligo. Participants were excluded if they reported any history of systemic diseases, such as autoimmune diseases, malignancy, hypertension, diabetes mellitus, and family history of autoimmune diseases. All patients were recruited from the dermatology clinic and had no systemic treatment four weeks before enrolment. Control subjects were healthy volunteers who were selected from physical examination center of the hospital during the same period, and volunteers with past or family history of vitiligo were excluded. Both the Ethics Committee of Affiliated Hospital of Xuzhou Medical University approval for the study (approval No. XYFY2021-KL252) and a written informed consent from all participants were obtained. This study complies with Declaration of Helsinki.
 |
Table 1 Primer Sequences |
Stable vitiligo is defined as static lesions of vitiligo without any new lesions or extension of previously existing lesions occurring over the past six months. By contrast, active vitiligo is defined as the recent appearance of new lesions or enlargement of existing lesions.25 A halo nevus is a benign melanocytic nevus that is surrounded by a hypopigmented zone.26
DNA Extraction
A total of 5 mL peripheral venous blood samples were drawn from each participant into EDTA-K2 anticoagulant tubes and stored at −80°C until DNA extraction. Genomic DNA was extracted from the whole blood by using a commercial Blood Genomic DNA preparation kit (Jizhen Biotechnology, Shanghai, China) following the manufacturer’s instructions.
Genotyping Analysis
Genotyping was carried out by the Snapshot sequencing technique. Polymerase chain reaction (PCR) was used for the amplification of target DNA sequences. The specific primer sequences are shown in Table 1.
A total of 15 μL of PCR reaction mixture was prepared in 10.1 μL of PCR water containing 25 mM MgCl 2 (1.5μL), 10 × PCR buffer (1.5 μL), 10 mM dNTPs (0.3 μL), l0 μM forward and reverse primer (0.15 μL each), Taq polymerase (0.3 μL), and sample DNA (1 μL). The PCR program was set under the following conditions: an initial denaturation temperature of 94°C for 3 minutes, followed by 30 cycles of 94°C for 15 seconds, 55°C for 15 seconds, 72°C for 30 seconds, and step of 72°C for 3 minutes. A total of 3 μL of PCR amplification product was then digested with 0.2 μL of Exonuclease I (ExoI) and 0.8 μL of Fast Alkaline Phosphatase (FastAP) and incubated at 37°C for 15 minutes and at 80°C for 15 minutes to remove the remaining primers and dNTPs. After purification of PCR products, the Snapshot multiplex single-base extension reaction was carried out in 6 μL solutions with 2 μL of the purified PCR products, 1 μL of Snapshot MIX (Applied Biosystems), 0.2 μL of extension reaction primer (10 μM), and 2.8 μL of PCR water. The extension primers were as follows: rs755622 (TAAGCCCGGCGCACCGCTCCAA); rs3747517 (TTTTTTTTTTTCGGAAATCATTAACTGTCTCA) ;rs1800796 (TTTTTTTTTTTTTTTTTTTTTTGCCAGGCAGTTCTACAACAGCC). The cycling conditions were 96°C for 1 minute, followed by 30 cycles of 96°C for 10 seconds, 52°C for 5 seconds, and 60°C for 30 seconds. Finally, extension products were further purified by ExoI and FastAP. The results of SNP typing were analyzed using an ABI 3730xl DNA Analyzer (Applied Biosystems) (Figure 1A–C).
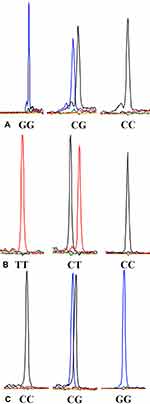 |
Figure 1 (A) Genotype sequencing of MIF-173G/C polymorphism; (B) Genotype sequencing of IFIH1 H843R polymorphism; (C) Genotype sequencing of IL6-572G/C polymorphism. |
Quantification of Serum Concentrations
Serum was obtained from all individuals at the time of inclusion; cytokine levels were quantified in a subset of 155 NSV patients and 117 control subjects. The determination of serum MIF, IFIH1 and IL6 concentrations was performed by ELISA kit (Jizhen Biotechnology, Shanghai, China), according to manufacturer’s protocol.
Statistical Analysis
Data management and analysis were performed using the Statistical Package for Social Sciences software (SPSS), version 23.0. For quantitative data, the mean, median, standard deviation (SD), and interquartile range (IQR) were calculated. Chi-square analysis was used to evaluate the Hardy–Weinberg equilibrium (HWE) and compare genotype and allele frequencies for each SNP between patients and controls. The differences in gender and age distribution between the two groups were analyzed using Chi-square test and Student’s t test, respectively. To compare nonparametric quantitative determinations, the Mann–Whitney U-test, Odds ratios (ORs) and 95% confidence intervals (CIs) were used to analyze the risk for NSV associated with the SNPs. A two-sided p value of less than 0.05 was considered statistically significant and 80% statistical power. The Bonferroni correction was applied to multiple tests, and p values were corrected accordingly to 0.0167 (adjust p value = alpha/n, alpha = 0.05, n = 3).
Results
Clinical Characteristics of the Study Population
There were no statistically significant differences in gender and age distribution among the vitiligo and control patient groups (p > 0.05). The demographic characteristics are summarized in Table 2. Both vitiligo patients and controls were in Hardy-Weinberg equilibrium with MIF-173G/C, IFIH1 H843R, and IL6-572G/C genotype distribution (p > 0.05) (Table 3).
 |
Table 2 Demographic Data Among the Patient Groups |
 |
Table 3 Hardy-Weinberg Balance Test Results of Non-Segmental Vitiligo and Control Group |
Association Between MIF-173G/C Polymorphism and Vitiligo
No significant differences in the genotype and allele frequencies of the MIF-173G/C polymorphism were found between patients with vitiligo and healthy controls (p > 0.0167) (Table 4). Notably, compared to the estimated sample size with PASS statistical software, the smaller sample size (155 patients and 117 controls) of this study decreased the statistical power significantly. Therefore, this conclusion we reached should be analyzed conservatively.
 |
Table 4 MIF-173G/C, IFIH1H843R, IL6-572G/C Genotypes and Allele Frequencies in Patient Groups |
Association Between IFIH1 H843R Polymorphism and Vitiligo
The genotype and allele frequencies of the IFIH1 H843R polymorphism among vitiligo cases and control subjects are shown in Table 4. There were significant differences in the distribution of the CT genotype between vitiligo patients and healthy controls (p = 0.030, OR = 1.766, 95% CI = 1.056–2.953). According to the dominance model, the carriers of the H843R C allele (CT + CC) showed an 1.847 fold risk (p = 0.013, OR = 1.847, CI = 1.136–3.004) to develop vitiligo than carriers of the T allele (TT). A significant difference in allele distribution was observed before and after Bonferroni correction between the two groups, and the C allele was present in 37.42% of patients and 27.35% of controls (p = 0.013).
Association Between IL6-572G/C Polymorphism and Vitiligo
The genotype and allele distribution of the IL6-572G/C polymorphism are summarized in Table 4. There were significant differences in the distribution of the CG and GG genotypes between vitiligo patients and healthy controls (p = 0.046, OR = 1.685, 95% CI = 1.009–2.816; p = 0.043, OR = 3.167, 95% CI = 0.988–10.145). According to the dominance model, the carriers of the −572 G allele (CG + GG) showed an 1.830-fold risk (p = 0.016, OR = 1.830, CI = 1.117–2.997) for the development of vitiligo than carriers of the −572 C allele (CC). A significant difference in allele distribution was observed before and after Bonferroni correction between the two groups, and the G allele was present in 29.03% of patients and 19.23% of controls (p = 0.009).
Associations Between the MIF-173G/C, IFIH1H843R, and IL6-572G/C Polymorphisms and Clinical Characteristics of Vitiligo
Stratified analysis of genotype, allele frequency, and clinical phenotype of vitiligo for the MIF-173G/C, IFIH1 H843R and IL6-572G/C polymorphisms are shown in Tables 5 and 6. The MIF-173G/C polymorphism in the active vitiligo group or with halo nevus vitiligo group was significantly higher than the stable vitiligo group or without halo nevus vitiligo group (p < 0.05), and the −173 C allele frequency in the active vitiligo group was significantly higher than the stable vitiligo group, which was statistically significant (p < 0.05). The IFIH1 H843R polymorphism in the early onset (≤ 20 years old), active or family history groups was significantly higher compared with the late onset (> 20 years old) vitiligo group, stable vitiligo group or no family history group (p < 0.05). The H843R C allele frequency in the early-onset vitiligo group, active vitiligo group or family history group was significantly higher than in the late-onset vitiligo group, stable vitiligo group or no family history group (p < 0.05). However, there was no correlation between the IL6-572G/C gene polymorphism and the clinical characteristics of patients with vitiligo (p > 0.05).
 |
Table 5 Distribution of MIF-173G/C, IFIH1 H843R, and IL6-572G/C Polymorphisms in Clinical Features of Vitiligo |
 |
Table 6 Allele Frequencies of MIF-173G/C, and IFIH1H843R Polymorphisms in Clinical Phenotypes of Vitiligo Patients |
MIF, IFIH1, IL6 Concentrations in Vitiligo Patients, Association with the MIF, IFIH1, IL6
We found differences in serum MIF, IFIH1, and IL6 concentrations between patients and controls. When we stratified the serum MIF, IFIH1 and IL6 concentrations according to the activity index of the patients, a higher MIF, IFIH1 and IL6 concentration were found in active patients in comparison with the stable ones [39.23 ng/mL (32.48–52.49) vs 37.89 ng/mL (32.57–40.71), p = 0.029; 38.61 ng/mL (34.66–45.68) vs 30.55 ng/mL (27.86–35.59), p < 0.001; 34.67 pg/mL (27.29–40.58) vs 27.67 pg/mL (21.18–30.68), p < 0.001]; and with the control groups [39.23 ng/mL (32.48–45.68) vs 24.78 ng/mL (13.89–31.27), p < 0.001; 38.61 ng/mL (34.66–45.68) vs 23.57 ng/mL (17.49–29.10), p < 0.001; 34.67 pg/mL (27.29–40.58) vs 19.78 pg/mL (12.57–27.48), p < 0.001] (Figure 2A–C).
Regarding MIF genotype analysis, active NSV patients with CC+CG genotypes exhibited significantly higher mean serum levels of MIF [37.58 ng/mL (31.01–52.13) vs 28.67 ng/mL (25.42–34.23), p = 0.002]. Regarding IFIH1 genotype analysis, active NSV patients with TT and CC+CT genotypes exhibited significantly higher mean serum levels of IFIH1 [38.61 ng/mL (34.66–45.37) vs 32.11 ng/mL (29.54–38.99), p = 0.017; 38.61 ng/mL (34.67–46.57) vs 32.11 ng/mL (29.54–38.99), p < 0.001]. Regarding IL6 genotype analysis, active NSV patients with CC and CG+GG genotypes exhibited significantly higher mean serum levels of IL6 [36.32 pg/mL (30.56–47.35) vs 27.68 pg/mL (23.59–34.54), p < 0.001; 34.64 pg/mL (23.59–38.45) vs 26.68 pg/mL (20.32–30.37), p = 0.011] (Figure 2D–F).
Discussion
Vitiligo is a complex, multifactorial, and polygenic disease with a multifaceted pathogenesis. It is well known that the interaction between genetics and environment influences the onset and evolution of vitiligo. Currently, more attention is paid to the factors that influence the autoimmune response.
MIF is a key proinflammatory cytokine that plays a role in inflammation, autoimmune, metabolic diseases, and cancer.27 In recent years, the MIFrs755622(−173G/C) polymorphism has been shown to be associated with the pathogenesis and progression of autoimmune diseases, such as rheumatoid arthritis (RA),28 psoriasis,29 systemic sclerosis (SSc),30 and systemic lupus (SLE).31 A study in a population of western Mexicans confirmed a significant association between MIF gene polymorphisms (−794CATT5-8 and −173G/C) and NSV.13 Due to resource and technical limitations of the present study, we focused on the MIF-173G/C gene polymorphism, and the results showed that there was no statistical difference in genotype and allele frequency of the MIF-173G/C polymorphism between vitiligo patients and the healthy control group (p > 0.05). This discrepancy between findings may be due to genetic heterogeneity in vitiligo susceptibility between ethnic populations or the small sample size involved in this study. Further analysis of the correlation between the MIF-173G/C allele distribution and clinical manifestations in patients with non-segmental vitiligo showed that it was more significant in patients with active vitiligo than in patients with stable vitiligo.
Evidence has shown that the occurrence of the MIF-173G/C polymorphism is related to its high transcriptional activity and protein expression. In vitro experiments have confirmed that −794CATT5-8 and −173 C significantly enhanced MIF gene transcription.32 The MIF-173 C allele has been associated with increased MIF levels in peripheral blood in a population-validated study.33 Multiple studies have found that the serum MIF concentration in patients with vitiligo vulgaris is significantly higher than the control group, and the serum MIF concentration in the active phase is higher compared with the stable phase.6,15,34–36 Ma et al37 and Garcia-Orozco et al13 further found that MIF mRNA levels in skin lesions of patients with vitiligo were significantly increased, and serum MIF concentration and in situ expression were correlated with active non-segmental vitiligo, further suggesting that MIF plays a role in the pathogenesis of non-segmental vitiligo, especially in active patients. In our study, we found significant differences between patients and controls in terms of serum MIF levels. Besides, serum MIF concentrations were not related to the presence of polymorphisms in patients with vitiligo. In conclusion, we found no association between MIF gene polymorphisms and the risk of NSV in Han population. However, further studies with a large sample are necessary to verify the association between NSV and the MIF gene polymorphisms.
IFIH1 encodes interferon-induced RNA helicases, which recognize the intracytoplasmic viral dsRNA, modulates interferon (IFN) responses and products of pro-inflammatory cytokines, including IL6, IL8, and TNF-α that have been proposed to play an essential role in the pathogenesis of vitiligo.36,38 In addition, IFIH1 plays a role as a virus-associated pattern recognition receptor (PRR) to activate innate immune responses and has a central role in triggering autoimmune responses, such as vitiligo.18 Some scholars have found that the activation of MDA5 could induce some chemokines of keratinocytes by using Poly (I : C) to simulate viral infection, thus exacerbating melanocyte death in vitiligo.39 Therefore, the MDA5 signaling pathway can be used as a potential therapeutic target for viral invasion of vitiligo.
In European populations, IFIH1rs2111485 was found to be associated with vitiligo by GWAS as a protective gene.18,19 The second GWAS in non-Hispanic European population confirmed that IFIH1rs1990760 and rs2111485 were susceptible loci of vitiligo, especially rs2111485.40 Similar results have been reported by Onan et al41 with reference to a Turkish population, further indicating that rs2111485 is a susceptible locus of vitiligo in Western populations. In the present study, rs3747517 (H843R), another SNP found in the IFIH1 gene, is a missense variant located at exon 13 of IFIH1.42 In humans, the variant of the IFIH1 gene that encodes the H843R polymorphism has been associated with different susceptibilities to multiple viral infections and autoimmune diseases, such as type I diabetes mellitus (T1DM),43 chronic viral hepatitis,44 systemic lupus erythematosus,45 and psoriatic arthritis.46 In the present study, significant associations were observed between IFIH1 H843R alleles and vitiligo susceptibility. Consistent results have been reported by Cheng et al20 in the Chinese Han population but did not specifically explore the genotype and allele frequency of this locus in relationship to the risk of vitiligo and its clinical impacts. In addition, we found that the IFIH1 H843R polymorphism genotype and allele frequency were higher in patients with early-onset (≤ 20), active or family history of vitiligo was significantly increased compared with late-onset (> 20), stable or no family history of vitiligo (p < 0.05). This may demonstrate differences in the genetic background of early-onset and late-onset vitiligo, active and stable vitiligo, with or without family history of vitiligo. To clarify, as definitively as possible, the contribution of IFIH1 promoter polymorphisms to vitiligo risk and phenotype in this study, we also determined the relationship between H843R polymorphisms and the serum levels of IFIH1. In the first instance, there were significant differences between patients and controls in terms of serum IFIH1 levels. Besides, serum IFIH1 concentrations were related to the presence of polymorphisms in patients with vitiligo.
IL6 is a well-known pro-inflammatory cytokine that has been implicated in vitiligo pathogenesis. Monocytes and macrophages are the major producers of IL6, but keratinocytes and melanocytes can also generate IL6 after stimulation. IL6 plays an important role in the human cytokine network, inducing the expression of various proteins responsible for acute inflammation and regulating endocrine and metabolism. IL6 is a paracrine inhibitor of human melanocyte proliferation and melanin production,47 and can significantly increase the expression of intercellular adhesion molecule-1 (ICAM-1) on the surface of melanocytes, which contributes to the adhesion between leukocytes and melanocytes, resulting in melanocyte damage and loss in patients with vitiligo.48 In addition, IL6 can induce the activation of polyclonal B cells, which may directly inhibit the growth of melanocytes and immune damage, leading to skin discoloration lesions.49 Besides, elevated IL6 expression in the skin and serum of vitiligo patients have been repeatedly identified,23,50,51 and the same results were found in our study.
There were significant racial differences between Asian and Caucasian populations in IL6 gene promoter region polymorphisms, and the allele C at the IL6-572G/C locus has been frequently observed in East Asian populations.52–54 Generally, the darker the skin color, the higher the probability of incidence of vitiligo. The difference between populations at two loci may be involved. The −572G/C is the most common polymorphic site in the IL6 promoter region, and we found that this site can result in an overproduction of IL6 and promote vitiligo development.55,56 Singh et al49 showed that the IL6-572G/C polymorphism was associated with vitiligo susceptibility in the Gujarat population and had a reduced risk of vitiligo in individuals with GC+CC genotypes compared with GG genotype (p < 0.05). Aydıngöz et al found no correlation between the IL6-174G/C polymorphism and vitiligo in Turkish population.57 In our study, we observed that the CG+GG genotype and G allele appeared more frequently among vitiligo patients, whereas the G allele carriers showed a 1.718-fold higher risk for vitiligo development compared with C allele carriers. In addition, we found no potential genetic association between IL6rs1800796 and clinical features in patients with vitiligo.
There were certain limitations in the current study. The smaller sample size in the investigation made the negative results less convincing. We did not further verify the correlation between variant genotypes and protein transcription levels due to the high cost of the experiment. Furthermore, we investigated only one polymorphism from each of the three genes and had a relatively small sample size. Therefore, confirming the genetic effects of multiple polymorphisms in MIF, IFIH1, and IL6 need to be further investigation.
Conclusion
The present study indicated that the IFIH1 H843R and IL6-572G/C polymorphisms play a role in NSV susceptibility in the Chinese population, and the significantly high prevalence of these two polymorphisms was observed in NSV. The H843R C allele may be associated with the incidence of vitiligo in youth (≤ 20 years old), active or family history groups. However, no statistical significance was observed between the MIF-173G/C polymorphism and NSV, which contradicts previous finding in other populations. The −173C allele may be associated with vitiligo incidence in the active NSV group in Chinese population. Moreover, the serum concentrations of MIF is associated with active NSV, and the serum IFIH1 and IL6 concentrations were related to the presence of polymorphisms in patients with NSV. Further investigation with a larger sample size and different ethnic groups is required to verify the results.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ezzedine K, Whitton M, Pinart M. Interventions for vitiligo. JAMA. 2016;316:1708–1709. doi:10.1001/jama.2016.12399
2. Hu DY, Ren YQ, Zhu KJ, et al. Comparisons of clinical features of HLA-DRB1*07 positive and negative vitiligo patients in Chinese Han population. J Eur Acad Dermatol Venereol. 2011;25:1299–1303. doi:10.1111/j.1468-3083.2010.03971.x
3. Birlea SA, Gowan K, Fain PR, et al. Genome-wide association study of generalized vitiligo in an isolated European founder population identifies SMOC2, in close proximity to IDDM8. J Invest Dermatol. 2010;130:798–803. doi:10.1038/jid.2009.347
4. Boniface K, Seneschal J, Picardo M, et al. Vitiligo: focus on clinical aspects, immunopathogenesis, and therapy. Clin Rev Allergy Immunol. 2018;54(1):52–67. doi:10.1007/s12016-017-8622-7
5. Sushama S, Dixit N, Gautam RK, et al. Cytokine profile (IL-2, IL6, IL-17, IL-22, and TNF-α) in vitiligo-New insight into pathogenesis of disease. J Cosmet Dermatol. 2019;18(1):337–341. doi:10.1111/jocd.12517
6. Song S, Xiao Z, Dekker FJ, et al. Macrophage migration inhibitory factor family proteins are multitasking cytokines in tissue injury. Cell Mol Life Sci. 2022;79(2):105. doi:10.1007/s00018-021-04038-8
7. Eldesouky F, Ibrahim AM, Sharaf SM. Macrophage migration inhibitory factor in alopecia areata and vitiligo: a case-controlled serological study. J Clin Aesthet Dermatol. 2020;13(10):24–27.
8. Feily A, Yaghoobi R, Pazyar N. Macrophage migration inhibitory factor as an incriminating agent in dermatological disorders. Indian J Dermatol. 2013;58(2):157. doi:10.4103/0019-5154.108068
9. Jankauskas SS, Wong DWL, Bucala R, et al. Evolving complexity of MIF signaling. Cell Signal. 2019;57:76–88. doi:10.1016/j.cellsig.2019.01.006
10. Zulu I, Hassan G, Njobvu RNL, et al. Cytokine activation is predictive of mortality in Zambian patients with AIDS-related diarrhoea. BMC Infect Dis. 2008;8:156. doi:10.1186/1471-2334-8-156
11. Gupta Y, Pasupuleti V, Du W, et al. Macrophage migration inhibitory factor secretion is induced by ionizing radiation and oxidative stress in cancer cells. PLoS One. 2016;11(1):e0146482. doi:10.1371/journal.pone.0146482
12. Moretti S, Fabbri P, Baroni G, et al. Keratinocyte dysfunction in vitiligo epidermis: cytokine microenvironment and correlation to keratinocyte apoptosis. Histol Histopathol. 2009;24(7):849–857. doi:10.14670/HH-24.849
13. Garcia-Orozco A, Martinez-Magaña IA, Riera-Leal A, et al. Macrophage inhibitory factor (MIF) gene polymorphisms are associated with disease susceptibility and with circulating MIF levels in active non-segmental vitiligo in patients from western Mexico. Mol Genet Genomic Med. 2020;8(10):e1416. doi:10.1002/mgg3.1416
14. Ahmad S, Mu X, Yang F, et al. Breaching self-tolerance to alu duplex RNA underlies MDA5-mediated inflammation. Cell. 2018;172(4):797–810.e13. doi:10.1016/j.cell.2017.12.016
15. Zhang S, Chu C, Wu Z, et al. IFIH1 contributes to M1 macrophage polarization in ARDS. Front Immunol. 2021;11:580838. doi:10.3389/fimmu.2020.580838
16. Yoneyama M, Kikuchi M, Matsumoto K, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. doi:10.4049/jimmunol.175.5.2851
17. Jin Y, Andersen GHL, Santorico SA, et al. Multiple functional variants of IFIH1, a gene involved in triggering innate immune responses, protect against vitiligo. J Invest Dermatol. 2017;137(2):522–524. doi:10.1016/j.jid.2016.09.021
18. Jin Y, Andersen G, Yorgov D, et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat Genet. 2016;48(11):1418–1424. doi:10.1038/ng.3680
19. Cheng L, Liang B, Tang XF, et al. Validation of susceptibility loci for vitiligo identified by GWAS in the Chinese Han population. Front Genet. 2020;11:542275. doi:10.3389/fgene.2020.542275
20. Xu Q, Fan D, Li F, et al. Influence of serum HMW adiponectin level in patients with pregnancy-induced hypertension syndrome on the occurrence of eclampsia in secondary pregnancy. Exp Ther Med. 2017;14(5):4972–4976. doi:10.3892/etm.2017.5112
21. Zailaie MZ. Decreased proinflammatory cytokine production by peripheral blood mononuclear cells from vitiligo patients following aspirin treatment. Saudi Med J. 2005;26(5):799–805.
22. Ascoli BM, Parisi MM, Bristot G, et al. Attenuated inflammatory response of monocyte-derived macrophage from patients with BD: a preliminary report. Int J Bipolar Disord. 2019;7(1):13. doi:10.1186/s40345-019-0148-x
23. Farhan J, Al-Shobaili HA, Zafar U, et al. Interleukin-6: a possible inflammatory link between vitiligo and type 1 diabetes. Br J Biomed Sci. 2014;71(4):151–157. doi:10.1080/09674845.2014.11669980
24. Ezzedine K, Lim HW, Suzuki T, et al. Revised classification/nomenclature of vitiligo and related issues: the vitiligo global issues consensus conference. Pigment Cell Melanoma Res. 2012;25(3):E1–13. doi:10.1111/j.1755-148X.2012.00997.x
25. Wang LM, Lu WJ, Yuan JT, et al. Utility of dermoscopy for evaluating the therapeutic efficacy of tacrolimus ointment plus 308-nm excimer laser combination therapy in localized vitiligo patients. Exp Ther Med. 2018;15(4):3981–3988. doi:10.3892/etm.2018.5911
26. Kim YY, Kim MY, Kim TY. Development of halo nevus around nevus spilus as a central nevus, and the concurrent vitiligo. Ann Dermatol. 2008;20(4):237–239. doi:10.5021/ad.2008.20.4.237
27. O’Reilly C, Doroudian M, Mawhinney L, et al. Targeting MIF in cancer: therapeutic strategies, current developments, and future opportunities. Med Res Rev. 2016;36(3):440–460. doi:10.1002/med.21385
28. Bae SC, Lee YH. Associations between circulating macrophage migration inhibitory factor (MIF) levels and rheumatoid arthritis, and between MIF gene polymorphisms and disease susceptibility: a meta-analysis. Postgrad Med J. 2018;94(1108):109–115. doi:10.1136/postgradmedj-2017-134934
29. Hernández-Bello J, Rodríguez-Puente M, Gutiérrez-Cuevas J, et al. Macrophage migration inhibitory factor gene polymorphisms (SNP −173 G>C and STR-794 CATT5-8) confer risk of plaque psoriasis: a case-control study. J Clin Lab Anal. 2021;35(11):e23999. doi:10.1002/jcla.23999
30. Baños-Hernández CJ, Navarro-Zarza JE, Bucala R, et al. Macrophage migration inhibitory factor polymorphisms are a potential susceptibility marker in systemic sclerosis from southern Mexican population: association with MIF mRNA expression and cytokine profile. Clin Rheumatol. 2019;38(6):1643–1654. doi:10.1007/s10067-019-04459-8
31. De la Cruz-Mosso U, Bucala R, Palafox-Sánchez CA, et al. Macrophage migration inhibitory factor: association of −794 CATT5-8 and −173 G>C polymorphisms with TNF-α in systemic lupus erythematosus. Hum Immunol. 2014;75(5):433–439. doi:10.1016/j.humimm.2014.02.014
32. Baugh JA, Chitnis S, Donnelly SC, et al. A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun. 2002;3(3):170–176. doi:10.1038/sj.gene.6363867
33. Radstake TR, Sweep FC, Welsing P, et al. Correlation of rheumatoid arthritis severity with the genetic functional variants and circulating levels of macrophage migration inhibitory factor. Arthritis Rheum. 2005;52(10):3020–3029. doi:10.1002/art.21285
34. Serarslan G, Yönden Z, Söğüt S, et al. Macrophage migration inhibitory factor in patients with vitiligo and relationship between duration and clinical type of disease. Clin Exp Dermatol. 2010;35(5):487–490. doi:10.1111/j.1365-2230.2009.03617.x
35. ElGhareeb MI, Mokadem SE, Sayed BE, et al. Soluble CD27 and MIF as possible serum biomarkers of vitiligo activity in Egyptian patients in Sharkia Governorate. Dermatol Rep. 2019;11(2):8265. doi:10.4081/dr.2019.8265
36. Farag AGA, Hammam MA, Habib MS, et al. Macrophage migration inhibitory factor as an incriminating agent in vitiligo. An Bras Dermatol. 2018;93(2):191–196. doi:10.1590/abd1806-4841.20186068
37. Ma L, Xue HB, Guan XH, et al. Relationship of macrophage migration inhibitory factor levels in PBMCs, lesional skin and serum with disease severity and activity in vitiligo vulgaris. Braz J Med Biol Res. 2013;46(5):460–464. doi:10.1590/S0100-879X2012007500152
38. Zhuang T, Yi X, Chen J, et al. Intracellular virus sensor MDA5 exacerbates vitiligo by inducing the secretion of chemokines in keratinocytes under virus invasion. Cell Death Dis. 2020;11(6):453. doi:10.1038/s41419-020-2665-z
39. Wang S, Liu D, Jin R, et al. Differential responses of normal human melanocytes to intra- and extracellular dsRNA. DNA Cell Biol. 2015;34(6):391–399. doi:10.1089/dna.2014.2711
40. Jin Y, Birlea SA, Fain PR, et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44(6):676–680. doi:10.1038/ng.2272
41. Onan D, Yorulmaz A, Ezgü FS, et al. The role of IFIH1 gene rs1990760 and rs2111485 single-nucleotide polymorphisms in generalized vitiligo predisposition. Turk J Med Sci. 2019;49(1):206–211. doi:10.3906/sag-1808-63
42. Mine K, Yoshikai Y, Takahashi H, et al. Genetic susceptibility of the host in virus-induced diabetes. Microorganisms. 2020;8(8):1133. doi:10.3390/microorganisms8081133
43. Yang H, Wang Z, Xu K, et al. IFIH1 gene polymorphisms in type 1 diabetes: genetic association analysis and genotype-phenotype correlation in Chinese Han population. Autoimmunity. 2012;45(3):226–232. doi:10.3109/08916934.2011.633134
44. Zhu XB, Zhuo LY, Yue M, et al. Genetic variants in IFIH1 and DDX58 influence hepatitis C virus clearance in Chinese Han population. J Med Virol. 2019;91(6):1097–1103. doi:10.1002/jmv.25398
45. Zhang J, Liu X, Meng Y, et al. Autoimmune disease associated IFIH1 single nucleotide polymorphism related with IL-18 serum levels in Chinese systemic lupus erythematosus patients. Sci Rep. 2018;8(1):9442. doi:10.1038/s41598-018-27782-7
46. Budu-Aggrey A, Bowes J, Stuart PE, et al. A rare coding allele in IFIH1 is protective for psoriatic arthritis. Ann Rheum Dis. 2017;76(7):1321–1324. doi:10.1136/annrheumdis-2016-210592
47. Moretti S, Spallanzani A, Amato L, et al. New insights into the pathogenesis of vitiligo: imbalance of epidermal cytokines at sites of lesions. Pigment Cell Res. 2002;15(2):87–92. doi:10.1034/j.1600-0749.2002.1o049.x
48. Kirnbauer R, Charvat B, Schauer E, et al. Modulation of intercellular adhesion molecule-1 expression on human melanocytes and melanoma cells: evidence for a regulatory role of IL6, IL-7, TNF beta, and UVB light. J Invest Dermatol. 1992;98(3):320–326. doi:10.1111/1523-1747.ep12499793
49. Morelli JG, Norris DA. Influence of inflammatory mediators and cytokines on human melanocyte function. J Invest Dermatol. 1993;100(2 Suppl):191S–195S. doi:10.1038/jid.1993.75
50. Ranjkesh MR, Partovi MR, Pashazadeh M. The study of serum level of Interleukin-2, Interleukin-6, and tumor necrosis factor-alpha in stable and progressive vitiligo patients from sina hospital in Tabriz, Iran. Indian J Dermatol. 2021;66(4):366–370. doi:10.4103/ijd.IJD_300_20
51. Singh S, Singh U, Pandey SS. Serum concentration of IL6, IL-2, TNF-α, and IFNγ in Vitiligo patients. Indian J Dermatol. 2012;57(1):12–14. doi:10.4103/0019-5154.92668
52. Wernstedt I, Eriksson AL, Berndtsson A, et al. A common polymorphism in the interleukin-6 gene promoter is associated with overweight. Int J Obes Relat Metab Disord. 2004;28(10):1272–1279. doi:10.1038/sj.ijo.0802763
53. Kou L, Yang N, Dong B, et al. Interaction between SELP genetic polymorphisms with inflammatory cytokine interleukin-6 (IL6) gene variants on cardiovascular disease in Chinese Han population. Mamm Genome. 2017;28(9–10):436–442. doi:10.1007/s00335-017-9712-9
54. Han SH, Lee NR, Kim HJ, et al. Association between the IL6, IL-10, and TNFα gene polymorphisms and preterm-birth in Korean women. Genes Genomics. 2020;42(7):743–750. doi:10.1007/s13258-020-00946-4
55. Zhao N, Liu HJ, Sun YY, et al. Role of interleukin-6 polymorphisms in the development of allergic rhinitis. Genet Mol Res. 2016;15(1):14–17.
56. Singh M, Jadeja SD, Vaishnav J, et al. Investigation of the role of interleukin 6 in vitiligo pathogenesis. Immunol Invest. 2022;51(1):120–137. doi:10.1080/08820139.2020.1813756
57. Aydıngöz IE, Kanmaz-özer M, Gedikbaşi A, et al. The combination of tumour necrosis factor-α −308A and interleukin-10-1082G gene polymorphisms and increased serum levels of related cytokines: susceptibility to vitiligo. Clin Exp Dermatol. 2015;40(1):71–77. doi:10.1111/ced.12446
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

