Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
Assessment of the Role of Selected SMAD3 and SMAD4 Genes Polymorphisms in the Development of Colorectal Cancer: Preliminary Research
Authors Wosiak A , Wodziński D , Michalska K , Pietrzak J , Kordek R, Balcerczak E
Received 14 September 2020
Accepted for publication 12 December 2020
Published 29 January 2021 Volume 2021:14 Pages 167—178
DOI https://doi.org/10.2147/PGPM.S281958
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Martin H Bluth
Agnieszka Wosiak,1 Damian Wodziński,1 Katarzyna Michalska,1 Jacek Pietrzak,1 Radzisław Kordek,2 Ewa Balcerczak1
1Laboratory of Molecular Diagnostics and Pharmacogenomics, Department of Pharmaceutical Biochemistry and Molecular Diagnostics, Interfaculty Cathedral of Laboratory and Molecular Diagnostics, Medical University of Lodz, Lodz 90-151, Poland; 2Department of Pathology, Cathedral of Oncology, Medical University of Lodz, Lodz 92-213, Poland
Correspondence: Ewa Balcerczak
Medical University of Lodz, Muszynskiego 1, Lodz 90-151, Poland
Tel/Fax +48 42 677 91 30
Email [email protected]
Background: Colon cancer is one of the most common types of malignant tumor worldwide. The molecular mechanism of colorectal carcinogenesis is very complex and not yet fully understood. The TGFβ (transforming growth factor β) signaling pathway plays a significant role in the development of many cancers, including colorectal cancer pathogenesis. Changes in TGFβ pathway are associated with increased colorectal cancer risk, because this pathway participates in the control of important cellular processes such as cell growth, proliferation, differentiation, or apoptosis. The family of SMAD (similar to mother against decapentaplegic) proteins is closely correlated to this pathway. SMADs genes expression affects modulation of the transcription of many genes, which leads to the inhibition of cell-growth and apoptosis in colon epithelial cells. The presence of SNPs (single nucleotide polymorphisms) in SMADs genes encoding proteins involved in the control of biological processes important for the cell may play a significant role in the predisposition to the development of colorectal cancer, or in the regulation of the severity of changes related to tumor growth. Extension of data in this field may provide clinically significant conclusions influencing the implementation of personalized treatment based on specific changes characteristic of a patient with colorectal cancer.
Purpose: The subject of this research was genotyping polymorphisms of SMAD3 (rs6494629) and SMAD4 (rs10502913, rs12968012, rs1057520801) genes in the group of patients with colorectal cancer and in the control group, and comparing the genotypic frequency distributions with clinical-pathological features within the study group and between the groups.
Materials and Methods: SNP genotyping analysis was performed on genomic DNA isolated from 84 frozen tissue sections of colorectal cancer and from 60 peripheral blood samples of patients without cancer. To evaluate the polymorphic variants of SMAD genes, the restricted fragment length of a polymorphism reaction (PCR-RFLP) was used.
Results: The results obtained in the study showed no significant association between the examined polymorphisms and the risk of developing colorectal cancer.
Conclusion: More extensive studies to confirm the results obtained in this study are needed. Further studies on a larger study group divided according to the clinical stage and histological differentiation may allow finding or excluding the significance of the studied SNPs as potential markers of colorectal cancer in relation to the clinico-pathological data.
Keywords: SMAD3 gene, SNP, colorectal cancer, SMAD4 gene
Introduction
Colorectal cancer (CRC) is a cancer commonly occurring around the world and it has a very high cancer-related mortality rate. In recent years morbidity of this malignancy has been significantly increased. The number of newly diagnosed cases of colon cancer at an advanced stage of the disease is still growing, especially in the developed countries. Therefore, CRC is an important clinical problem in healthcare.1,2 One of the research directions which may contribute to reducing the mortality rate in patients with colorectal cancer is expanding knowledge about the biological and molecular path leading to the development of this cancer. Molecular pathogenesis of CRC is heterogeneous and comprise a range of genomic and epigenomic alterations.3,4
One of the molecular subtypes (CMS4) of colorectal cancer is characterized by the changes of TGFβ pathway that play an important role in the pathogenesis of many cancers development, being engaged in the regulation of many diverse biological processes within cells of the gastrointestinal epithelium. The TGFβ pathway is responsible for the control of critical cellular processes involved in carcinogenesis, such as cell growth, proliferation, differentiation, or apoptosis. Additionally, this pathway is associated with angiogenesis and inflammation.5–7 Activation of the TGFβ cascade protects normal epithelial cells from excessive proliferation and differentiation, and is responsible for controlling programmed cell death, but also has a negative effect on cell adhesion or migration.6–8 The TGFβ path plays a dual role in carcinogenesis, at the stage of cancer initiation and the early progression of the TGFβ trail inhibits cell proliferation and the directs cell to the apoptosis; during cancer progression its protective role is reduced.8–10
In the TGFβ signaling pathway, the ligand binds to the TGFβ II receptor on the cell surface that phosphorylates the TGFβ1 receptor. The latter one activates SMAD2 and SMAD3 proteins and forms a heterodimer complex with SMAD4, which regulates transcription factors in the cell nucleus, such as FOXL24, SMIF5, or TGIF8.11,12 Disorders such as loss of chromosomal 18q and other aberrations leading to abolition of signal transmission through the TGFβ pathway may contribute to a transformation of normal colonic cells to cancerous cells. Loss of heterozygosity in 18q21 where SMAD4 gene is located, occurs in 60–70% of colorectal cancer cases.13 In the case of cancers, the TGFβ/SMAD signaling pathway may act as a tumor promoter, leading to uncontrolled proliferation of cancer cells.9 Therefore, research to identify TGFβ path-dependent mechanisms is needed, for example into the mechanisms of TGFβ signaling in conjunction with SMAD4.
The family of SMAD proteins is closely-related to TGF-β signaling, as central mediators in this pathway. This group of proteins is responsible for the regulation of cellular responses by transducing signals from the cell membrane to nucleus and affecting the role in maintaining tissue homeostasis. SMADs proteins can be divided into three distinct classes: R-SMADs (the so-called receptor regulated SMADs), Co-SMADs (common-mediator SMADs, including representative SMAD4) and I-SMADs (inhibitory SMADs). SMAD4 protein is a critical component of TGFβ signaling. The SMAD4 gene, encoding the SMAD4 protein, acts as a tumor suppressor gene and probably prevents the development of various cancers, mainly pancreatic, breast, and colorectal cancer.14,15 Homozygous deletions or intragenic mutations, leading to SMAD4 gene inactivation or loss of their functions, were detected with higher frequency in the late stages of these carcinomas. This dependence indicates the participation of the SMAD4 gene in tumor initiating, development, or progression.16,17 The SMAD3 gene, located on the long arm of the 15th chromosome, encodes the protein mediating intracellular signal transmission. The SMAD3 gene also plays a key role in TGFβ/SMAD modulation of transcription of many genes, including genes encoded pro-inflammatory cytokines, for example CDKI (cyclin dependent kinase inhibitor) genes functioning as tumor suppressor genes. In this mechanism, SMAD3 gene expression is crucial for cell-growth inhibition and apoptosis of various types of epithelial cells including the colon.18–20 SMADs genes belong to highly polymorphic genes, and mutations in these genes are observed frequently in cases of colorectal cancer.21
The main aim of this research was an assessment of impact of the selected SMAD3 and SMAD4 genes polymorphisms in CRC development and progress. In this study, genotyping for polymorphisms at positions: rs10502913, rs12968012, rs1057520801, of the SMAD4 gene and rs6494629 of SMAD3 gene was done for better understanding of SNPs influence on pathomechanisms, where the investigated genes play an important role. Much data has been published including the assessment of abnormalities in the TGFβ/SMAD pathway in the development of colorectal cancer. Based on the confirmed role of this pathway in the pathogenesis of colorectal cancer, we tried to expand the available knowledge with data on the analyzed SNPs. There are few reports in the literature on the importance of the studied SNPs in colorectal cancer, while there are only reports on the importance of the studied SNPs in other diseases, such as gastric cancer or lung cancer. The subject is important because there are no data on the relationship between the studied genotypes and alleles and the development and progression of colorectal cancer, especially in the case of high-stage cancer in the Polish population.
Materials and Methods
Materials
Study tissue specimens were received during tumor surgeries from patients diagnosed with colorectal cancer on the basis of histological examination. All collected tissue samples of colorectal carcinomas were immediately deeply frozen in liquid nitrogen and stored at −80°C temperature until DNA isolation was performed.
The study group consisted of 84 cases of colorectal cancer diagnosed at the Department of Pathology of the Medical University of Lodz. The characteristic of study group in terms of demographic and clinical–pathological features is presented in Table 1.
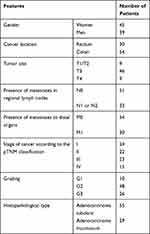 |
Table 1 Study Group Characteristics |
Blood samples from the control group of patients were also stored at −20°C until further analysis. The control group consisted of 60 blood samples obtained from healthy individuals from the local blood bank.
Both groups were ethnically and geographically matched. All experiments on these biological materials were carried out in accordance with the principles of the Declaration of Helsinki and were approved by the Ethical Committee of the Medical University of Lodz RNN/82/20/KE and KE/227/20. All subjects included in the study gave their informed consent.
Methods
DNA Extraction
Evaluation of polymorphic variants of the SMAD genes was performed on genomic DNA isolated from frozen tissue sections of colorectal cancer and from peripheral blood samples of patients without cancer.
Genomic DNA was isolated from biological materials using a column method and the Genomic Midi Kit (A&A Biotechnology, Poland) in accordance with the manufacturer’s protocols. The concentration and purity of DNA obtained after isolation was assessed spectrophotometrically. DNA samples that had the ratio of the absorbance at 260/280 nm between of 1.8–2.0 were selected for further study.
Genotyping
DNA obtained from tissues and peripheral blood isolation was used to determine single nucleotide polymorphisms of the SMAD genes. The evaluation of SMAD3 gene rs6494629 C/T polymorphism and SMAD4 gene rs1057520801 A/C, rs10502913 G/A, and rs12968012 C/G polymorphisms, respectively, was carried out using the restriction fragment length polymorphism reaction (PCR-RFLP). SMAD genes fragments, including the polymorphic sites, were amplified using a specific pair of primers. Amplification product sizes and primers sequences are shown in Table 2. PCR reaction for all specific DNA fragments was performed in a total reaction volume of 20 μL containing: 50 ng of genomic DNA, 3.5 μL of 10× PCR buffer, 0.4 μL of 0.2 mM each deoxynucleotide triphosphate, 0.7 μL of 25 mM MgCl2, 0.5 μL of 10 µmol of forward and reverse primers and 0.2 μL of 2.5 U/µL of JumpStart™ DNA polymerase and distilled water up to 20 μL. PCR parameters for SMAD3 gene rs6494629 C/T polymorphism were as follows: an initial denaturation at 94°C for 2 minutes, followed by 34 cycles including 94°C for 15 seconds (denaturation), 64°C for 30 seconds (annealing), 72°C for 30 seconds (extension) and an additional extension step at 72°C for 5 minutes. PCR reaction conditions for the amplification of SMAD4 gene fragments included 95°C for 2 minutes, 34 cycles covering 94°C for 30 seconds, 59°C for 30 seconds, 72°C for 60 seconds, and final step at 72°C for 5 minutes. Negative control (without DNA) was included in every experiment. PCR products were evaluated during electrophoresis in 2% agarose gel stained with ethidium bromide and viewed under UV light. Products of PCR amplification (the presence of a band of the right size in electrophoresis) were subjected to digestion with specific restriction enzymes. The digested products were electrophoresed on 2% agarose gel. Genotypes for analyzed polymorphisms were assessed based on different banding patterns. The presence of specific bands in electrophoresis for individual genotypes is presented in Table 3.
 |
Table 2 Amplification Product Sizes and Sequences of Primers |
 |
Table 3 Banding Patterns for Genotypes |
Statistical Analysis
STATISTICA 13.3 statistical software (TIBCO 2020) was used for data analysis. The Chi-squared Pearson test was applied to evaluate the conformity between the observed and expected genotype frequencies in the investigated and control groups.
Differences in genotype frequencies among colorectal cancer patients and control group, and the association of the various genotypes with clinical parameters were determined using the Chi-squared Pearson test, Chi-squared with Yates’correction test, V-squared test, and Fisher’s exact test. For all statistical comparisons the P-values<0.05 were considered as statistically significant.
Results
All samples from the study and control group were successfully analyzed. For all studied SNPs the observed genotype frequency distributions in both groups were consistent with the Hardy–Weinberg equilibrium. The Chi-squared test P-values for selected SNPs are presented in Table 4.
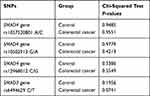 |
Table 4 Genotype Frequency Distributions (Hardy–Weinberg Equilibrium) |
Firstly, the frequency of genotype distributions of selected SNPs between colorectal cancer patients and healthy controls were compared and results are presented in Table 5. The differences in the frequency of individual genotypes in groups were not at the level of statistical significance. This comparison suggests that polymorphic variants of investigated genes probably do not predispose to an increased individual risk of developing colorectal cancer.
 |
Table 5 Frequency of Genotype Distributions of Selected SNPs Between the Control and Investigated Group |
Statistical analysis of the genotype frequency distribution for SMAD4 gene rs1057520801 A/C was not carried out, because in the study group only one case with the AC genotype was observed with the other cases having the AA genotype.
The distribution of genotype variants of the SMAD4 polymorphisms was compared to demographic features such as gender and family history of colorectal cancer. The results are shown in Table 6. Despite the fact that AA genotype (for SMAD4 gene rs10502913 G/A) and GG genotype (for SMAD4 gene rs12968012 C/G) were more commonly seen in women than in men and in people with a positive family history of colorectal cancer, no statistically significant differences were observed.
 |
Table 6 The Distribution of Genotype Variants of the SMAD4 Polymorphisms Depending on the Demographic Features |
Then, genetic variants of the SMAD polymorphisms were compared with clinical and pathological parameters. The obtained results are shown in Tables 7 and 8.
 |
Table 7 The Distribution of Genotype Variants of the SMAD4 Polymorphisms Depending on the Clinical-Pathological Features |
 |
Table 8 The Distribution of Genotype Variants of the SMAD3 Polymorphism Depending on the Demographic and Clinical-Pathological Features |
The group of patients with colorectal cancer was divided into four subgroups according to the pTNM classification: less advanced stage pTNM defined as I and II and more advanced stage as III and IV. No significant differences between the examined genotypes and pTNM were found.
The next comparable parameter was histological grading. In this case, the group of patients with colorectal cancer was divided into three subgroups G1, G2, and G3 according to the grade of the tumor malignancy. No significant association between genotype distributions among patients with different histological grading was observed, despite the GG genotype (for SMAD4 gene rs12968012 C/G) and TT genotype (for SMAD3 gene rs6494629 C/T) were the most common in the G1 subgroup of patients and the GG genotype (for SMAD4 gene rs10502913 G/A) was the least frequent in G1 group.
In further analysis the studied genotype distribution with clinical-pathological features like tumor localization, tumor size, and histopathological type was compared. No significant differences were found, although the GG genotype (for SMAD4 rs12968012 C/G) was more frequently observed in colon cases than in rectum cases.
For the analysis of the presence of metastases in regional lymph nodes, patients with colorectal cancer were again divided into three groups: without regional nodes involvement by cancer and with cancer involved regional nodes (one or more). No statistically significant dependence was observed in this case.
In the next step of the statistical analysis the genotype distribution of SNPs SMAD genes were compared to other clinical-pathological features, like the presence of tumor-infiltrating lymphocytes which is a beneficial prognostic factor in advanced colon cancer and infiltration of blood vessels by tumor. Also in this case there were no statistically significant differences.
Haplotype analysis of SMAD4 gene polymorphisms was performed within the study and control groups. An assessment of correlation between the occurrence of a specific SMAD4 haplotype variants and demographic or clinical features within study group was performed. No statistically significant correlations were observed. The P-values for individual analyses are shown in Tables 5–7.
Discussion
In this study the association between SMAD3 rs6494629 C/T, SMAD4 rs1057520801 A/C, rs10502913 G/A, rs12968012 C/G genes polymorphisms, and the risk of incidence and progression of colorectal cancer were evaluated. The analysis was carried out on DNA samples isolated from colorectal cancer tissues and on DNA samples isolated from peripheral blood obtained from healthy individuals. All analyses were performed within the European population and the obtained genotype frequencies of the SNPs remain consistent with the population data provided by the dbGaP database. The genotype frequency distribution of SNPs in SMAD3 and SMAD4 genes between the group of patients with colorectal cancer and the control group was analyzed. The frequencies of particular genotypes in the two groups did not differ significantly. Then, we compared the distribution of genotype variants with clinic-pathological features within the study group; however, no statistically significant differences were observed. The lack of association between the distribution of genotype variants of the analyzed single nucleotide polymorphisms and the explored parameters may be caused by small aberrations within the study group in the case of cancer invasion and metastasis. The examined group of patients with colorectal cancer included more patients with an early or initial stage of cancer compared to the number of patients with its advanced stage. To have a full view of frequency genotype distribution of SMAD3 and SMAD4 SNPs in different stages of colon cancer development, research on a group of patients with more advanced colon cancer should be performed.
Many polymorphisms of the SMAD3 gene in pathogenesis of various disease entities have been described; for example, in the study by Zhang et al,20 an association between rs12901499 polymorphism and osteoarthritis in the Chinese population was confirmed.
Until now the rs6494629 C/T polymorphism has not been evaluated in cases of colon cancer. In the publication written by Paradowska-Gorycka et al,22 the authors examined rs6494629 C/T SNP in the SMAD3 gene and they found the association with susceptibility to the development of rheumatoid arthritis. They observed that the occurrence of the TT genotype was significantly higher in the group of patients with rheumatoid arthritis compared to the group of healthy people. In addition, they noticed a significant correlation between the occurrence of SMAD3 rs6494629 T allelic variant and the severity of disease symptoms. The authors showed that SMAD3 gene polymorphisms may lead to irregularities in the course of SMAD signaling pathway and assumed a hypothesis that the examined polymorphism may influence the changes in the regulation of T cell response, because the SMAD3 gene is involved in this process. The presence of the SNP rs6494629 genotype TT could lead to severity of rheumatoid arthritis symptoms.
Based on the assumptions that the SMAD3 gene is involved in the regulation of the immune response, the analysis was carried out on a study group including patients with diagnosed colorectal cancer. Chronic intestinal inflammation belongs to predisposing conditions for the development of colon cancer. The chronic inflammatory process of the large intestine promotes the neoplastic transformation of normal intestinal epithelial cells. The risk of developing colon cancer associated with inflammation in patients with inflammatory bowel disease such as Crohn's disease or ulcerative colitis is much higher.2 In this study, the association between rs6494629 C/T genotypes frequencies and predisposition to the development of colon cancer was examined and no correlation was observed. Further studies are necessary on more clinical materials derived from patients with colon cancer at different stages.
A common change in colorectal cancer pathogenesis is the loss of DNA sequence from the long arm of chromosome 18q21, within which chromosome region, the SMAD4 gene is located. The loss of heterozygosity on chromosome 18 is observed in approximately 70% of colorectal cancer cases, with the loss of the SMAD4 gene locus in about 30–40% of patients. This may indicate an association between the loss of function of this gene and the development of colorectal cancer.23 Despite this fact, many publications suggest the role of the SMAD4 gene at advanced stages of cancer. Reduction or loss of SMAD4 gene expression is a frequent feature of cancer in the large intestine, although this event has been more common during colorectal cancer progression in patients with distant metastases of cancer.24,25 Research results in this regard suggest that mutations play a significant role in mechanisms of SMAD4 gene inactivation in colorectal carcinogenesis.26
Associations between the occurrence of germline mutations in the SMAD4 gene and susceptibility to colorectal cancer are known. Therefore, the involvement of somatic mutations in this gene may be associated with a predisposition to the development of sporadic colon cancers.26 The SMAD4 gene is highly polymorphic, thus the participation of genetic variants of this gene may play a particular role in the risk of colorectal cancer. Extensive data based on the results obtained with genome-wide association studies provide information on many SNPs of various genes that may be correlated with the genetic predisposition to develop colorectal cancer.27,28 This report is one of a few published studies in which the role of SMAD4 polymorphisms in colorectal cancer was assessed.
In the research conducted by Slattery et al,12 the influence of genetic variations in the genes involved in the TGFβ pathway on the risk of developing colon and rectal cancer was assessed. The researchers observed a correlation between the polymorphic variant rs10502913 of the SMAD4 gene and an increased risk of rectal cancer in men, and a decreased risk of rectal cancer in women.
Wu et al21 aimed at searching for the association between selected SMAD4 gene polymorphisms and gastric cancer risk, and obtained significant conclusions. Four single nucleotide polymorphisms located within the introns of the SMAD4 gene have been chosen for the study, the rs10502913 variant of the SMAD4 gene was assessed, among others. The results obtained in this analysis proved that two variants of genotypes for selected SNPs were associated with a reduced risk of gastric cancer and could play a protective role in developing of cancer in a Chinese population. The authors also observed a correlation between the occurrence of these genotypic variants and a higher level of SMAD4 gene expression.
The subject of this research is important because there are no data on the correlation of the studied genotypes and alleles with the development and progression of colorectal cancer, especially with high clinical stage in the Polish population. Based on the current knowledge, a hypothesis was drawn about the possible association of the studied SNPs with colon cancer. Preliminary studies have not shown a significant association, which provides new knowledge about the potential utility of selected SNPs as genetic markers in colorectal cancer.
Conclusion
More extensive studies to confirm the results obtained in this study are needed. Further studies on a larger study group divided according to the clinical stage and histological differentiation may allow finding or excluding the significance of the studied SNPs as potential markers of colorectal cancer in relation to the clinico-pathological data.
Disclosure
The authors declare that there is no conflict of interest in this work.
References
1. Kamocki ZK, Wodynska NA, Zurawska JL, Zareba KP. Significance of selected morphological and histopathological parameters of colon tumors as prognostic factors of cancer spread. Turk J Gastroenterol. 2017;28(4):248–253. doi:10.5152/tjg.2017.16734
2. Jacobs D, Zhu R, Luo J, et al. Defining early-onset colon and rectal cancers. Front Oncol. 2018;8:504. doi:10.3389/fonc.2018.00504
3. Sameer AS. Colorectal cancer: molecular mutations and polymorphisms. Front Oncol. 2013;3:114. doi:10.3389/fonc.2013.00114
4. Druliner BR, Ruan X, Sicotte H, et al. Early genetic aberrations in patients with sporadic colorectal cancer. Mol Carcinog. 2018;57(1):114–124. doi:10.1002/mc.22738
5. Wideł MS, Wideł M. Mechanizmy przerzutowania i molekularne markery progresji nowotworów złośliwych. I. Rak jelita grubego. Postepy Hig Med Dosw. 2006;60:453–470.
6. Tian M, Schiemann WP. The TGF-beta paradox in human cancer: an update. Future Oncol. 2009;5:259–271. doi:10.2217/14796694.5.2.259
7. Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov. 2012;11:790–811.
8. Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi:10.1126/science.1071809
9. Slattery ML, Herrick JS, Lundgreen A, Wolff RK. Genetic variation in the TGF-β-signaling pathway and colon and rectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2011;20(1):57–69. doi:10.1158/1055-9965.EPI-10-0843
10. Li Q, Wu H, Chen B, et al. SNPs in the TGF-βsignaling pathway are associated with increased risk of brain metastasis in patients with non-small-cell lung cancer. PLoS One. 2012;7(12):e51713. doi:10.1371/journal.pone.0051713
11. Schmierer B, Hill CS. TGFβ–SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8(12):970. doi:10.1038/nrm2297
12. Slattery ML, Lundgreen A, Herrick JS, et al. Associations between genetic variation in RUNX1, RUNX2, RUNX3, MAPK1 and eIF4E and risk of colon and rectal cancer: additional support for a TGF-β-signaling pathway. Carcinogenesis. 2011;32(3):318–326. doi:10.1093/carcin/bgq245
13. Woodford-Richens KL, Rowan AJ, Gorman P, et al. SMAD4 mutations in colorectal cancer probably occur before chromosomal instability, but after divergence of the microsatellite instability pathway. Proc Natl Acad Sci USA. 2001;98(17):9719–9723. doi:10.1073/pnas.171321498
14. Fleming NI, Jorissen RN, Mouradov D, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73(2):725–735. doi:10.1158/0008-5472.CAN-12-2706
15. Du X, Pan Z, Li Q, Liu H, Li Q. SMAD4 feedback regulates the canonical TGF-β signaling pathway to control granulosa cell apoptosis. Cell Death Dis. 2018;9(2):151. doi:10.1038/s41419-017-0205-2
16. Salovaara R, Roth S, Loukola A, et al. Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut. 2002;51(1):56–59. doi:10.1136/gut.51.1.56
17. Sarshekeh AM, Advani S, Overman MJ, et al. Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS One. 2017;12(3):e0173345. doi:10.1371/journal.pone.0173345
18. Mithani SK, Balch GC, Shiou S-R, Whitehead RH, Datta PK, Beauchamp RD. Smad3 has a critical role in TGF-β-mediated growth inhibition and apoptosis in colonic epithelial cells. J Surg Res. 2004;117(2):296–305. doi:10.1016/S0022-4804(03)00335-4
19. van de Laar IM, van der Linde D, Oei EH, et al. Phenotypic spectrum of the SMAD3-related aneurysms-osteoarthritis syndrome. J Med Genet. 2012;49:47–57. doi:10.1136/jmedgenet-2011-100382
20. Zhang L, Zhang H, Wang W, Zhao Y. Association between SMAD3 gene rs12901499 polymorphism and knee osteoarthritis in a Chinese population. J Clin Lab Anal. 2018;32:e22383. doi:10.1002/jcla.22383
21. Wu DM, Zhu HX, Zhao QH, et al. Genetic variations in the SMAD4 gene and gastric cancer susceptibility. World J Gastroenterol. 2010;16(44):5635–5641. doi:10.3748/wjg.v16.i44.5635
22. Paradowska-Gorycka A, Romanowska-Prochnicka K, Haladyj E, Manczak M, Maslinski S, Olesinska M. Association of the Smad3 and NFATc2 gene polymorphisms and their serum levels with susceptibility to rheumatoid arthritis in Polish cohorts. Clin Exp Immunol. 2015;179(3):444–453. doi:10.1111/cei.12482
23. Maitra A, Molberg K, Albores-Saavedra J, et al. Loss of Dpc4 expression in colonic adenocarcinomas correlates with the presence of metastatic disease. Am J Pathol. 2000;157(4):1105–1111. doi:10.1016/S0002-9440(10)64625-1
24. Xie W, Rimm DL, Lin Y, et al. Loss of smad signaling in human colorectal cancer is associated with advanced disease and poor prognosis. Cancer J. 2003;9(4):302–312. doi:10.1097/00130404-200307000-00013
25. Alazzouzi H, Alhopuro P, Salovaara R, et al. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res. 2005;11(7):2606–2611.
26. Miyaki M, Iijima T, Konishi M, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18(20):3098–3103. doi:10.1038/sj.onc.1202642
27. Samani NJ, Erdmann J, Hall AS, et al. Genome wide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi:10.1056/NEJMoa072366
28. Doaei S, Hajiesmaeil M, Aminifard A, et al. Effects of gene polymorphisms of metabolic enzymes on the association between red and processed meat consumption and the development of colon cancer; a literature review. J Nutr Sci. 2018;7(26):1–7. doi:10.1017/jns.2018.17
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
