Back to Journals » International Journal of General Medicine » Volume 10
Assessment of sputum smear-positive but culture-negative results among newly diagnosed pulmonary tuberculosis patients in Tanzania
Authors Mnyambwa NP , Ngadaya ES , Kimaro G, Kim D, Kazwala R, Petrucka P , Mfinanga SG
Received 19 March 2017
Accepted for publication 15 June 2017
Published 12 July 2017 Volume 2017:10 Pages 199—205
DOI https://doi.org/10.2147/IJGM.S137469
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Nicholaus Peter Mnyambwa,1,2 Esther S Ngadaya,2 Godfather Kimaro,2 Dong-Jin Kim,1 Rudovick Kazwala,3 Pammla Petrucka,1,4 Sayoki G Mfinanga2
1School of Life Sciences and Bioengineering, Nelson Mandela African Institution of Science and Technology, Arusha, Tanzania; 2National Institute for Medical Research, Muhimbili Medical Research Center, Dar es Salaam, Tanzania; 3Department of Veterinary Medicine, Sokoine University of Agriculture, Morogoro, Tanzania; 4College of Nursing, University of Saskatchewan, Saskatoon, Canada
Abstract: Diagnosis of pulmonary tuberculosis (TB) in technology-limited countries is widely achieved by smear microscopy, which has limited sensitivity and specificity. The frequency and clinical implication of smear-positive but culture-negative among presumptive TB patients remains unclear. A cross-sectional substudy was conducted which aimed to identify the proportion of nontuberculous mycobacteria (NTM) infections among 94 “smear-positive culture-negative” patients diagnosed between January 2013 and June 2016 in selected health facilities in Tanzania. Out of 94 sputa, 25 (26.60%) were GeneXpert® mycobacteria TB positive and 11/94 (11.70%) repeat-culture positive; 5 were Capilia TB-Neo positive and confirmed by GenoType MTBC to be Mycobacterium tuberculosis/Mycobacterium canettii. The remaining 6 Capilia TB-Neo negative samples were genotyped by GenoType® CM/AS, identifying 3 (3.19%) NTM, 2 Gram positive bacteria, and 1 isolate testing negative, together, making a total of 6/94 (6.38%) confirmed false smear-positives. Twenty-eight (29.79%) were confirmed TB cases, while 60 (63.83%) remained unconfirmed cases. Out of 6 (6.38%) patients who were HIV positive, 2 patients were possibly coinfected with mycobacteria. The isolation of NTM and other bacteria among smear-positive culture-negative samples and the presence of over two third of unconfirmed TB cases emphasize the need of both advanced differential TB diagnostic techniques and good clinical laboratory practices to avoid unnecessary administration of anti-TB drugs.
Keywords: nontuberculous mycobacteria, pulmonary TB, smear-positive, culture-negative, tuberculosis diagnosis
Introduction
In 2014, the World Health Organization (WHO) adopted a new strategy to reduce tuberculosis (TB) deaths by 90% and TB incidence by 80% within the 15 years window from 2015 to 2030; however, diagnosis of TB remains challenging,1 particularly in resource-constrained countries and in the contexts of high TB incidence rates. The widely used simple and inexpensive sputum smear microscopy for diagnosis of pulmonary TB2,3 suffers from both lack of sensitivity and specificity.3,4 The performance of smear microscopy is further reduced in patients with extrapulmonary TB, those coinfected with HIV,4,5 and those with nontuberculous mycobacteria (NTM).5,6
NTM are ubiquitous bacteria found in water and soil7,8and are well known to cause lung diseases.9,10 In recent years, pulmonary NTM infection rates have dramatically increased worldwide7,8,11 and widely vary across geographical scale and human population. Defining the clinical implication of NTM infection in settings with endemic TB, like Tanzania, requires discrimination of NTM from Mycobacterium tuberculosis (MTB) complex suspects.12 Shared morphological characteristics between NTM and the MTB complex contributes to the poor performance of smear microscopy.8 Thus, NTM infections are commonly misdiagnosed as pulmonary TB, particularly in resources-limited countries.7,8 Despite these limitations, sputum smear microscopy remains a mainstay technique for clinical diagnosis of pulmonary TB in low and middle-income countries and remains an integral part of the global TB control strategy.6 Current WHO TB guidelines recommend immediate initiation of anti-TB drugs and a regular follow-up schedule after smear-positive results. Thus, the limited capacity of smear examination to differentiate MTB complex from NTM infection is not taken into account, resulting in unnecessary people undergoing TB treatment.
Mycobacterial culture is more sensitive and highly specific than smear examination, and is hence considered the gold standard for diagnosis of TB.13 Identification of species from culture is based on phenotypic and biochemical tests which are not definitive,9 and nonviable bacterial cells also result in culture-negative.14 NTM infections can falsely present as culture positive and can even be misdiagnosed as multidrug-resistant TB.7,15 The clinical implication of culture false-positive has remained elusive. Nucleic acid amplification assays like GenoType® CM/AS (Hain Lifescience, Nehren, Germany) and GeneXpert® MTB/RIF (Cepheid, Sunnyvale, CA, USA) can be applied for quick identification and confirmation of TB, though the techniques are expensive for routine diagnosis. This study used both phenotypic and molecular techniques to identify the likelihood of NTM or other TB-like infection among smear-positive culture-negative samples.
Materials and methods
Study design
This cross-sectional study, nested in the East Africa Public Health Laboratory Network (EAPHLN) project, utilized sputum specimens collected between February 2013 and June 2016. The broad objective of the EAPHLN project was to establish a network of efficient, high-quality, accessible public health laboratories for the diagnosis and surveillance of TB and other communicable diseases in East Africa.
Settings
The EAPHLN project collected samples from the following selected sites: Kibong’oto Infectious Diseases Hospital (in Kilimanjaro Region), Musoma Regional Referral Hospital (in Mara Region), Mnazi Mmoja Referral Hospital (in Zanzibar), Levolosi Health Centre (in Arusha Region), Nyamagana Health Centre (in Mwanza Region), St Vicent Health Centre in Mkuranga (in Pwani Region), and Shinyanga Regional Referral Hospital (in Shinyanga Region). During the period February 2013–June 2016, a total of 530 patients aged 15 years and above were diagnosed as smear positive. Eighteen percent (94/530) were “smear-positive and culture-negative.” Laboratory analysis to identify the proportion of NTM species among presumptive TB cases performed at the Central TB Reference Laboratory (CTRL), unless otherwise stated. CTRL is the only reference laboratory in the country, located in Dar es Salaam, that serves health facilities in all regions of the country. CTRL performs smear microscopy and microbial culture, participates in epidemiological research on TB, and evaluates and provides diagnostic services for the quality assurance of the National Tuberculosis Control Programme. As a part of quality assurance and laboratory strengthening, CTRL corroborates with a number of supranational TB laboratories like UK NEQAS, Uganda Supranational Laboratory as well as the Supranational Laboratory of the Antwerp Institute of Tropical Medicine, Belgium.
HIV testing
HIV tests were performed at the study site level as per standard care as specified in the national HIV and testing algorithm16 in order to determine the HIV status of the consented participants.
Smear examination
Sputum sample collection, processing, transportation, and detection of acid-fast bacilli (AFB) by Zeihl-Neelsen (ZN) were performed according to the National es for TB management.17 Smear-positive sputum samples were sent to CTRL for smear reexamination and culture. Laboratory scientists/technicians read the smears blindly and graded the results using the WHO system.
TB detection by GeneXpert MTB/RIF
Using unprocessed sputa samples, GeneXpert tests (Cepheid) were performed to all 94 sputa in accordance with the manufacturer’s instructions for detection of TB. GeneXpert MTB/RIF assay is an automated hemi-nested real-time polymerase chain reaction (PCR) assay for detection of both M. tuberculosis complex (MTBC) and rifampicin-resistant organisms in 2 hours. It amplifies an MTB-specific sequence of the 81-bp rpoB gene, which is probed with molecular beacons for mutations within the rifampicin-resistance determining region.
Repeat culture
Sputum samples from each study site were processed, packaged, and transported to CTRL, with a transit time of 2–3 days. At CTRL, samples were homogenized (using N-acetyl-cysteine) to free the bacilli from the mucus, cells, or tissue, and this was followed by decontamination procedure (using NaOH) that liquefies the organic debris and eliminates the unwanted normal flora. Processed sediments were used to inoculate Löwenstein -Jensen (LJ) culture media according to standard operating procedure. Repeat culture was performed after completion of sample collection period, using sputum specimens that were stored at –80°C after initial culture. The inoculated culture was incubated and observed every week for 8 weeks before being regarded as negative. A culture was considered as contaminated following observation of overgrowth of microorganisms that were lacking characteristics of mycobacteria. Repeat cultures were performed to all 94 smear-positive but culture-negative sputa. Reexamination with ZN stain was performed to confirm the presence of AFB among NTM suspects.
Species identification
The Capilia TB-Neo assay (an immunochromatographic test) was performed to all repeat-culture positive samples in accordance with the manufacturer’s instructions (TAUNS Laboratories, Inc., Shizuoka, Japan) to differentiate MTB complex from NTM infections. The Capilia TB-Neo detects MPT64 protein secreted by species of MTB complex. Isolates of MTB complex detected by Capilia TB-Neo were further subjected to a GenoType MTBC assay (Hain Lifescience, Nehren, Germany) for confirmation and identification to the species level. In order to identify species that were Capilia TB-Neo negative (NTM suspects), 2 additional genotyping methods: the GenoType CM/AS (Hain Lifescience) were performed according to the manufacturer’s instructions. GenoType CM can simultaneously detect and differentiate up to 27 clinically relevant NTM species from MTBC. GenoType AS can detect and differentiate an additional 19 NTM species.
Demographic and clinical data
Patients’ demographic variables (age and sex) and clinical information (HIV status and TB treatment history) were extracted from the EAPHLN project register books and tools at the sites and CTRL.
Definitions
As stated earlier, a definitive diagnosis of pulmonary TB is difficult to achieve; however, based on this study, we establish that:
- True TB smear-positive (confirmed TB infection) = smear positive and confirmation by any of the nucleic acid amplification tests (GeneXpert, or GenoType CM/AS or GenoType MTBC).
- False smear-positive (confirmed not TB infection) = smear positive, but nucleic acid amplification confirmed NTM or other AFB infection.
- Unconfirmed TB case = smear positive but negative in all other tests (culture and nucleic acid amplification).
Ethical consideration
The study was nested within the EAPHLN project which was approved by the National Health Research Ethics Sub-Committee of the National Institute for Medical Research, Tanzania. All study participants aged 18 years and above provided a written informed consent, and for those who were younger than 18 years, a written informed consent was obtained from parents or guardians.
Results
A total of 94 (17.74%) smear-positive culture-negative samples of presumptive TB patients who presented with TB symptoms from a pool of 530 smear-positive samples collected between January 2013 and June 2016 were analyzed. The remaining 436 (82.26%) of 530 samples were smear-positive culture-positive and so were not included in this analysis.
The overall mean age (SD) of the 94 patients was 40.54 (15.63) years. The majority, 54/94 (57.45%), of the participants were males with a mean (SD) age of 41.8 (17.04) years, and for female participants (40, 42.55%) the mean (SD) age was 38.85 (13.54) years. Table 1 reports the distribution of the participants by study sites
  | Table 1 Distribution of participants by study site |
Out of 94 sputum samples, 25 (26.60%) were GeneXpert positive (no rifampicin resistance detected), of which 2 were culture positive. A total of 69 (73.40%) sputa were GeneXpert negative, of which only 9 were culture positive. Only 2/25 GeneXpert positive samples were also culture positive.
Repeat LJ culture yielded 11 (11.70%) growths, 2 were from a pool of 25 GeneXpert positive samples and the remaining 9 from 69 GeneXpert negative sputa. Additionally, 2 culture growths were discarded due to contamination during culture preparation. Eighty-three (88.30%) remained negative on culture to the termination point that was after 8 eeks.
Of the 11 culture-positive samples, 5 tested Capilia TB-Neo positive for MTB complex and were confirmed by GenoType MTBC test as being MTB/Mycobacterium canettii. GenoType MTBC assay cannot distinguish MTB from M. canettii. The remaining 6 Capilia TB-Neo negative isolates were typed by GenoType CM, which identified 3/94 (3.19%) as NTM infections (2 Mycobacterium fortuitum, 1 Mycobacterium abscessus), 2 as Gram positive bacteria, and 1 isolate was negative. The 2 Gram positive bacteria and the negative isolate were subjected to GenoType AS assay, and all 3 were negative. Sixty (63.83%) sputa were negative in both GeneXpert and culture. Laboratory workflow and outcomes are summarized in Figure 1. Six (6.38%) of the 94 patients were HIV positive. Of the 2 M. fortuitum infected patients, 1 patient also had HIV, and 1 patient from a pool of 5 M. tuberculosis/M. canettii cases had also HIV coinfection.
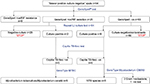  | Figure 1 Flowchart showing laboratory procedure and outcomes. Abbreviations: NTM, nontuberculous mycobacteria; TB, tuberculosis; LJ, Löwenstein–Jensen; RIF, rifampicin. |
Based on the tests performed and case definition established earlier, a total of 28/94 (29.79%) patients sputa (GeneXpert positive plus GenoType MTBC positives) were referred to as “confirmed TB cases” or “true smear-positive”, while 6 (6.38%) cases were referred to as “confirmed not TB infection” or “false smear-positive”; among these 3 were NTM infections, 2 were Gram positive bacteria, and 1 was not identified by the tests. The remaining 60 (63.83%) samples that were negative in both GeneXpert and culture were referred to as “unconfirmed cases.”
Discussion
Early detection and effective treatment are central components for successful management and control of every infectious disease including pulmonary TB. The diagnosis of pulmonary TB by sputum smear microscopy is seriously obstructed by its limited sensitivity and specificity.2,4 Although low sensitivity in the examination of stained acid-fast smears has been given high priority, little consideration is given to false smear-positive as it pertains to specificity.
Despite the fact that the sensitivity of smear microscopy is inferior to both GeneXpert and mycobacterial culture, this study reports that 60/94 smear-positive sputa were negative in both GeneXpert and culture. Due to the retrospective nature of this study, we failed to confirm the actual cause of these cases. However, laboratory technical failures and presence of nonviable bacilli can result into no growth in culture.18 Toxicity of reagents used for decontamination, centrifugation, transportation, and storage is all known to affect the viability of the bacilli if not performed properly.4 For similar reasons, decontamination procedure and centrifugation, if not accurately performed, might have contributed to the miss of 5/11 cases of M. tuberculosis/M. canettii on the initial culture but identified by the repeat culture. Demonstration studies documented high sensitivity and specificity (97%–100%) of GeneXpert to detect TB and RIF resistance; however, the technology requires stable electricity, annual recalibration, and compliance with temperature ceiling.19 The impact of these on the sensitivity of the instrument for routine diagnosis of pulmonary TB in places where there is a shortage and unstable power supply, like Tanzania, is not well established. However, due to the high specificity of this nucleic acid amplification technology, we concluded that 25 (26.60%) GeneXpert positive cases (2 confirmed by GenoType CM) were referred to as “true pulmonary TB cases.”
Recently, there has been increasing trend of respiratory infection worldwide due to NTM, which significantly varies across different geographical localities.7,8,12 However, older adults and immunocompromised persons like HIV-infected persons are at more risk of the infection.10,20 NTM infections present similar clinical and radiographic manifestations like those of pulmonary TB,7,21 hence resulting false in positives in both smear and culture. In Tanzania, it is commonly assumed that patients presenting with pulmonary infection are infected with MTB,12,22,23 with the possibility of other respiratory infection like NTM infection not taken into consideration. In this study, 3 patients (3.19%) were confirmed as having NTM infections (2 M. fortuitum and M. abscessus), but they may had been mistreated as pulmonary TB, which may result in worsening patient’s health, repeated visits to health facilities, and an increase in the risk of drug-resistance. NTM infection does not respond to anti-TB agents; however, the clinical impact of the indiscriminate use of anti-TB drugs in the tremendously, recently emerging drug-resistant TB era remains unassessed.2,24 Infection due to species of M. fortuitum and M. abscessus, which both have the capacity to cause respiratory infections in human, has previously been reported in Tanzania.25,26 Although this study did not conduct drug sensitivity testing, both M. fortuitum and M. abscessus can be misdiagnosed as resistant TB/multidrug-resistant TB due to their natural resistant pattern to antituberculosis drugs,15,26 which may result in unnecessary administration of toxic second-line anti-TB treatment. The prevalence of NTM reported in the present study, which seems to be lower than recently reported in the country,12,22 is not conclusive because of the limited sample size and because it only constitutes a small proportion of pulmonary TB diagnosis scenarios. Moreover, due to the retrospective nature of this study, we could not certainly implicate the isolated NTM as the cause (and the only cause) of the infection in these patients.
Additionally, various particles that are acid-fast (eg, food particles, dye precipitates, and non-mycobacterial species) can cause false smear-positive results. For example, Nocardia spp. and Rhodococcus equi20 have a similar appearance to that of mycobacterium on smear examination and can be misdiagnosed as pulmonary TB. Recently, one study in Tanzania isolated different species of Nocardia among TB suspects, suggesting that Nocardia spp. are an important cause of pulmonary infections that are merely underdiagnosed and/or ignored.27 Furthermore, poor quality of smear microscopy and reading errors may result in either failure to detect active TB or reporting of non-TB cases.28,29 In this study, 3/11 isolates (2 Gram positive bacteria and 1 negative isolate) could not be identified to the species level due to the limitations of the typing methods used in this study. GenoType CM can detect Gram positive bacteria with a high G+C content; however, carefully evaluation is needed to rule out the possibility of coexistence with mycobacterial species.30
Limitations
This study failed to assess the association of NTM infection with related chronic diseases (eg, HIV) and sociodemographic characteristic like age and sex due to the small size. It was also difficult to assess treatment outcomes among patients due to the retrospective nature of the study. Furthermore, this study could not rule out the possibility of bacterial colonization and/or contamination among the reported NTM or bacterial cases.
Conclusion
Our study reports that, out of 94 smear-positive cases, 64% were both GeneXpert and culture negative and identified nontuberculous mycobacteria among TB suspects. Less consideration of false smear-positive samples may result in mismanagement of a health condition. It is important that the national TB program considers strengthening the quality assurance system and tracking the performance of both smear microscopy and mycobacterial culture system regularly. Use of advanced differential TB diagnostic techniques and reviewing and emphasizing better clinical practices that ensure accuracy in diagnosis would help to avoid putting patients’ health and lives at risk.
Acknowledgments
This work was mainly supported by the EAPHLN project, which was appraised in February 2010 and implemented regionally within the East African countries with a soft loan from the World Bank. This work was partly supported by the DELTAS Africa Initiative [Afrique One-ASPIRE /DEL-15-008]. Afrique One-ASPIRE is funded by a consortium of donors including the African Academy of Sciences (AAS), Alliance for Accelerating Excellence in Science in Africa (AESA), the New Partnership for Africa’s Development Planning and Coordinating (NEPAD) Agency, the Wellcome Trust [107753/A/15/Z], and the UK government. The authors extend sincere thanks to the Nelson Mandela African Institution of Science and Technology for providing adequate learning resources and invaluable support and to the management of the CTRL where this piece of work was carried out. The views expressed in this publication are those of the authors and not necessarily those of Afrique One-ASPIRE or the World Bank.
Disclosure
The authors report no conflicts of interest in this work.
Reference
World Health Organization (WHO). Global Tuberculosis Report 2016. World Health Organization; 2016. [cited February 20, 2017]. Available from http://www.who.int/tb/publications/global_report/archive/en/ | ||
Pai M, Schito M. Tuberculosis diagnostics in 2015: landscape, priorities, needs, and prospects. J Infect Dis. 2015;211 (Suppl 2):S21–S28. | ||
McNerney R, Daley P. Towards a point-of-care test for active tuberculosis: obstacles and opportunities. Nat Rev Microbiol. 2011;9(3):204–213. | ||
Parsons LM, Somoskövi A, Gutierrez C, et al. Laboratory diagnosis of tuberculosis in resource-poor Countries: challenges and opportunities. Clin Microbiol Rev. 2011;24(2):314–350. | ||
Weyer K, Mirzayev F, Migliori GB, et al. Rapid molecular TB diagnosis: Evidence, policy making and global implementation of Xpert MTB/RIF. Eur Respir J. 2013;42(1):252–271. | ||
Ngabonziza JC, Ssengooba W, Mutua F, et al. Diagnostic performance of smear microscopy and incremental yield of Xpert in detection of pulmonary tuberculosis in Rwanda. BMC Infect Dis. 2016;16(1):660. | ||
Maiga M, Siddiqui S, Diallo S, et al. Failure to recognize nontuberculous mycobacteria leads to misdiagnosis of chronic pulmonary tuberculosis. PLoS One. 2012;7(5):e36902. | ||
Raju RM, Raju SM, Zhao Y, Rubin EJ. Leveraging advances in tuberculosis diagnosis and treatment to address nontuberculous mycobacterial disease. Emerg Infect Dis. 2016;22(3):365–369. | ||
Johnson MM, Odell JA. Nontuberculous mycobacterial pulmonary infections. J Thorac Dis. 2014;6(3):210–220. | ||
Henkle E, Aksamit T, Barker A, et al. Patient-centered research priorities for pulmonary Nontuberculous Mycobacteria (NTM) Infection. An NTM Research Consortium Workshop Report. Ann Am Thorac Soc. 2016;13(9):S379–S384. | ||
Kendall B, Winthrop K. Update on the epidemiology of pulmonary nontuberculous mycobacterial infections. Semin Respir Crit Care Med. 2013;34(1):87–94. | ||
Hoza AS, Mfinanga SGM, Rodloff AC, Moser I, König B. Increased isolation of nontuberculous mycobacteria among TB suspects in Northeastern, Tanzania: public health and diagnostic implications for control programmes. BMC Res Notes. 2016;9(1):109. | ||
Agrawal M, Bajaj A, Bhatia V, Dutt S. Comparative study of GeneXpert with ZN stain and culture in samples of suspected pulmonary tuberculosis. J Clin Diagn Res. 2016;10(5):DC09–DC12. | ||
Ryu YJ. Diagnosis of pulmonary tuberculosis: recent advances and diagnostic algorithms. Tuberc Respir Dis (Seoul). 2015;78(2):64–71. | ||
Shahraki AH, Heidarieh P, Bostanabad SZ, et al. Multidrug-resistant tuberculosis may be nontuberculous mycobacteria. Eur J Intern Med. 2015;26(4):279–284. | ||
National Aids Control Program (NACP). The United Republic of Tanzania. National Comprehensive Guidelines for HIV Testing and Counselling; 2013. [cited February 27, 2017]. Available from: http://www.nacp.go.tz/site/publications/hiv-counselling-and-esting. | ||
National Tuberculosis and Leprosy Program (NTLP). United Republic of Tanzania Ministry of Health and Social Welfare National Tuberculosis and Leprosy Programme Manual for the Management of Tuberculosis and Leprosy; 2015 [cited March 5, 2017]. Available from: http://ntlp.go.tz/index.php?option=com_phocadownload&view=sections&Itemid=139. | ||
Lee JS, Kim EC, Joo SI, et al. The incidence and clinical implication of sputum with positive acid-fast bacilli smear but negative in mycobacterial culture in a tertiary referral hospital in South Korea. J Korean Med Sci. 2008;23(5):767. | ||
Trébucq A, Enarson DA, Chiang CY, et al. Xpert ® MTB/RIF for national tuberculosis programmes in low-income countries: when, where and how? Int J Tuberc Lung Dis. 2011;15(12):1567–1571. | ||
Hatta M, Sultan AR, Tandirogang N, Masjudi, Yadi. Detection and identification of mycobacteria in sputum from suspected tuberculosis patients. BMC Res Notes. 2010;3(1):72. | ||
Koh WJ, Yu CM, Suh GY, et al. Pulmonary TB and NTM lung disease: comparison of characteristics in patients with AFB smear-positive sputum. Available from http://www.ingentaconnect.com/content/iuatld/ijtld/2006/00000010/00000009/art00011. Accessed March 18, 2017. | ||
Kilale AM, Ngadaya E, Muhumuza J, et al. Who has mycobacterial disease? A cross sectional study in agropastoral communities in Tanzania. PLoS One. 2016;11(5):e0153711. | ||
Hoza AS, Lupindu AM, Mfinanga SGM, Moser I, König B. The role of nontuberculous mycobacteria in the diagnosis, management and quantifying risks of tuberculosis in Tanga, Tanzania. Tanzan J Health Res. 18(2). Available from: http://www.ajol.info/index.php/thrb/article/view/120757. Accessed March 14, 2017. | ||
Mnyambwa NP, Kim DJ, Ngadaya ES, Kazwala R, Petrucka P, Mfinanga SG. Clinical implication of novel drug resistance-conferring mutations in resistant tuberculosis. Eur J Clin Microbiol Infect Dis. 2017:1–8. | ||
Mfinanga SG, Warren RM, Kazwala R, et al. Genetic profile of Mycobacterium tuberculosis and treatment outcomes in human pulmonary tuberculosis in Tanzania. Tanzan J Health Res. 2014;16(2). | ||
Mpagama SG, Heysell SK, Ndusilo ND, et al. Diagnosis and interim treatment outcomes from the first cohort of multidrug-resistant tuberculosis patients in tanzania. PLoS One. 2013;8(5):e62034. | ||
Hoza AS, Mfinanga SGS, Moser I, K?nig B. Isolation, biochemical and molecular identification of Nocardia species among TB suspects in northeastern, Tanzania; a forgotten or neglected threat? BMC Infect Dis. 2017;17(1):407. | ||
Manalebh A, Demissie M, Mekonnen D, Abera B, Kumar P. The quality of sputum smear microscopy in public-private mix directly observed treatment laboratories in West Amhara Region, Ethiopia. PLoS One. 2015;10(4):e0123749. | ||
Mfinanga GS, Ngadaya E, Mtandu R, et al. The quality of sputum smear microscopy diagnosis of pulmonary tuberculosis in Dar es Salaam, Tanzania. 2008. Available from: https://tspace.library.utoronto.ca/handle/1807/40824. Accessed March 14, 2017. | ||
Richter E, Rüsch-Gerdes S, Hillemann D. Evaluation of the GenoType Mycobacterium Assay for identification of mycobacterial species from cultures. J Clin Microbiol. 2006;44(5):1769–1775. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
