Back to Journals » Cancer Management and Research » Volume 13
Assessment of Potential Prognostic Value of Peroxiredoxin 1 in Oral Squamous Cell Carcinoma
Authors Shen Y , Xu H, Li L, Lu Y, Zhang M , Huang X, Tang X
Received 10 May 2021
Accepted for publication 28 June 2021
Published 15 July 2021 Volume 2021:13 Pages 5725—5737
DOI https://doi.org/10.2147/CMAR.S319048
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Yajun Shen, 1 Haoyue Xu, 2 Lingyu Li, 1 Yunping Lu, 1 Min Zhang, 1 Xin Huang, 2 Xiaofei Tang 1
1Beijing Institute of Dental Research, Beijing Key Laboratory, Beijing Stomatological Hospital & School of Stomatology, Capital Medical University, Beijing, 100050, People’s Republic of China; 2Department of Oral and Maxillofacial Surgery, Beijing Stomatological Hospital & School of Stomatology, Capital Medical University, Beijing, 100050, People’s Republic of China
Correspondence: Xiaofei Tang
Division of Oral Pathology, Beijing Institute of Dental Research, Beijing Stomatological Hospital & School of Stomatology, Capital Medical University, No. 4 Tiantanxili, Dongcheng District, Beijing, 100050, People’s Republic of China
Tel +86 01057099311
Fax +86 01057099310
Email [email protected]
Xin Huang
Department of Oral and Maxillofacial Surgery, Beijing Stomatological Hospital & School of Stomatology, Capital Medical University, No. 4 Tiantan Xili, Dongcheng District, Beijing, 100050, People’s Republic of China
Email [email protected]
Purpose: The role of the peroxiredoxin (PRDX) family in oral squamous cell carcinoma (OSCC) remains unclear. This study aimed to investigate the expression of PRDXs and their effects on the prognosis in OSCC.
Methods: The expression of PRDXs and their effects on prognosis were analysed in 216 OSCC samples from The Cancer Genome Atlas (TCGA) database. OSCC tissues and adjacent noncancerous tissues (ANTs) were obtained from 68 clinical patients. Quantitative real-time (qRT)-PCR, Western blot, and immunohistochemical (IHC) staining were used to verify the relationship between the expression level of PRDX1 and different clinical features. Gene set enrichment analysis (GSEA) was used to examine the molecular mechanism of PRDX1 in OSCC.
Results: PRDX1 was found to be the only gene in PRDX family that highly expressed in OSCC samples and affected the prognosis of patients with OSCC. PRDX1 expression was significantly related to tumor stage, lymphatic metastasis, and pathological grade. A nomogram consisting of tumor stage, N stage, and PRDX1 level was constructed. GSEA showed that high expression of PRDX1 involved many cancer-related molecular functions and signaling pathways.
Conclusion: PRDX1 may play an important role in the occurrence and development of OSCC, and may be a potential new target for OSCC treatment.
Keywords: peroxiredoxin 1, oral squamous cell carcinoma, lymph node metastasis, prognosis, nomogram
Corrigendum for this paper has been published
Further corrigendum has been published
Introduction
Head and neck cancer (HNC) is the seventh most common tumor with a high mortality rate worldwide.1 Oral squamous cell carcinoma (OSCC) accounts for approximately 95% of all HNCs and affects approximately 400,000 people every year.2 Although significant advances in surgery and other adjuvant treatments have greatly improved OSCC outcome, the 5-year survival rate of patients with OSCC remains less than 60%.3 An important risk factor contributing to the high mortality in OSCC is lymphatic metastasis, and the incidence of lymphatic metastasis in patients with OSCC is approximately 30%.4 The molecular mechanisms of tumor metastasis in OSCC remain unclear and the therapeutic targets are still unknown, making it difficult to prevent and treat metastasis in OSCC.5 Therefore, further evaluation of OSCC biomarkers is of great significance.
An imbalance between the production of reactive oxygen species (ROS) and their elimination through protective mechanisms can lead to oxidative stress.6 Somatic mutations may occur under continuous stimulation by peroxides that may further promote tumor formation.7 Interestingly, although oxidative stress promotes tumors, increased ROS level also presents a significant challenge to the survival of tumor cells.8 Redox homeostasis is also extremely important in tumor development.9 Due to the increased metabolism in tumors, tumor cells produce more ROS, however, elevated ROS clearance rates have been observed in many tumors and cancer cell lines to counteract the slightly higher levels of ROS that exist under normal physiological conditions.10
The redox homeostasis in tumors depends on endogenous antioxidants.11 Peroxiredoxins (PRDXs) are one of many enzymatic antioxidant systems present in different organelles. They use the thioredoxin (Trx)/Trx reductase/NADPH system as a reducing equivalent to remove H2O2 and promote or inhibit tumorigenesis by regulating the level of ROS, depending on the type of cancer. To date, a total of six isoforms have been identified in the PRDX family including PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, and PRDX6.12 PRDXs are distributed in various locations in cells; PRDX1, 2, and 6 are mainly located in the cytoplasm and nucleus, PRDX3 is expressed only in the mitochondria, PRDX4 is expressed in the endoplasmic reticulum, and PRDX5 is expressed in the cytoplasm and mitochondria.13 Several studies have found that PRDXs play important roles in redox homeostasis, cell differentiation, proliferation and apoptosis in several human tumor types, including lung, hepatocellular, breast, prostate and colon.14–18 some PRDXs also promote cell invasion and metastasis in many malignant tumors, high expression of PRDX2, PRDX5 and PRDX6 all promote metastasis of colorectal cancer.19–21 PRDX3 is associated with metastasis in uveal melanoma.22 Additionally, Increasing number of studies have found that PRDXs affect the prognosis of patients with various types of tumors, including breast, liver, ovarian, and gastric cancers.23–26 PRDX1, PRDX2, and PRDX6 have also been reported to be highly expressed in OSCC.27–29 To date, few studies have investigated the relationship between the PRDX family and metastasis and prognosis of OSCC.
In this study, we analyzed the expression of PRDX family members in 216 patients with OSCC from The Cancer Genome Atlas (TCGA) database. We found that, among the PRDX family members, PRDX1 showed the strongest impact on the prognosis of patients with OSCC, and was also significantly related to lymphatic metastasis, tumor stage, and pathological grade, suggesting that PRDX1 may potentially serve as an effective prognostic biomarker for patients with OSCC. Finally, we established a prognostic model of OSCC based on PRDX1. We further confirmed these results using 68 pairs of OSCC tissues and adjacent noncancerous tissues (ANTs).
Materials and Methods
Data Acquisition and Processing
The mRNA expression data and the corresponding clinical data of OSCC samples were obtained from the TCGA database. After deleting samples with low gene expression and those with incomplete clinical data, a total of 216 OSCC samples and 22 normal samples were obtained. A total of 68 pairs of OSCC tissues and ANTs were also collected from Beijing Stomatological Hospital of Capital Medical University. This research complied with the Declaration of Helsinki and was approved by the Research Ethics Committee of Beijing Stomatological Hospital of Capital Medical University (Approval No. CMUSH-IRB-KJ-PJ-2018-01). Patients were selected according to the following criteria: (1) patients pathologically diagnosed with OSCC, with no history of tumors in any other part; (2) patients with OSCC who underwent primary tumor resection and neck lymphatic dissection but did not receive previous adjuvant therapy such as radiotherapy and/or chemotherapy; and (3) patients with complete follow-up data. The tissues obtained during the operation were immediately frozen in liquid nitrogen for storage until quantitative real-time PCR and Western blot analysis. As all the patients were in M0 stage, patients in N0 stage were defined as the non-metastatic group, and patients in N1, N2, and N3 stages were defined as the metastatic group.
Transcriptional Levels of the PRDX Family Members
For normalization, the ‘apply’ function in R studio was performed to convert the counts data of all genes expression into transcripts per million (TPM) format for all the samples in TCGA cohort, and the mRNA expression values of PRDX1, PRDX2, PRDX3, PRDX4, PRDX5 and PRDX6 were extracted for further analysis. The expression differences of these six genes were compared separately between normal group and tumor group, as well as between metastatic group and non-metastatic group.
Prognostic Analysis of Patients with OSCC
To explore the relationship between expression levels of PRDXs and overall survival rate of patients with OSCC, the samples in the TCGA cohort were divided into high and low expression groups based on the median mRNA expression value of each PRDX. Kaplan-Meier (KM) analysis was performed for survival curve and the significance was determined by Log rank test. In order to further evaluate the prognostic hazard ratio and 95% confidence intervals of each PRDX in patients with OSSC, PRDX1, PRDX2, PRDX3, PRDX4, PRDX5 and PRDX6 were included in univariate Cox proportional-hazards regression analysis.
Western Blot Analysis
Proteins from 68 pairs of OSCC tissues and ANTs were extracted using radioimmunoprecipitation assay (RIPA) lysis buffer containing a proteinase inhibitor cocktail (Sigma, USA). Total protein concentration was determined using the Bradford method. Equal amounts of total protein from each sample were loaded onto a 10% gel for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% skimmed milk for 1 h at 25°C, and then incubated with primary antibodies (rabbit anti-PRDX1, 1:2000 dilution, rabbit anti-HSP90, 1:5000 dilution, Abcam, USA) at 4 °C overnight. Next, the membranes were incubated with secondary antibodies: 1:2000 dilution (Amersham Biosciences, USA) for 1h and visualized using enhanced chemiluminescence reagent (Bio-Rad, USA).
Quantitative Real-Time Transcription (qRT)-PCR
Total RNA was extracted from 68 pairs of OSCC tissues and ANTs using TRIzol (Invitrogen Life Technologies, USA) reagent according to the manufacturer’s instructions. cDNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA). PRDX1 and GAPDH expression levels were determined by qRT-PCR using SYBR Green (Qiagen, Germany). The sequences for the primers used were as follows: PRDX1 (forward 5ʹ-GGGTATTCTTCGGCAGATCA-3ʹ, reverse 5ʹ-TCCCCATGTTTGTCAGTGAA-3ʹ), and GAPDH (Forward 5ʹ-AGGTCGGTGTGAACGGATTTG-3ʹ, reverse 5ʹ-TGTAGACCATGTAGT TGAGGTCA-3ʹ). Gene expression was calculated using the 2−ΔΔCT method.
Immunohistochemical (IHC) Staining
IHC staining was performed to detect the expression of PRDX1 in different pathological grades of tissues from the 68 pairs of OSCC tissues and ANTs. The tissues were fixed in formalin and embedded in paraffin. After antigen retrieval, all the tissue sections were blocked with goat serum, and then incubated with rabbit anti-PRDX1 (1:1000 dilution, Abcam, USA) primary antibody at 4 °C overnight. Next, the sections were incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (MaiXin, China) at 37 °C for 30 min. Finally, diaminobenzidine tetrahydrochloride (DAB; MaiXin, China) was used as a chromogenic substrate for staining, and hematoxylin was used for counterstaining.
The sections were evaluated under a microscope (Olympus, Tokyo, Japan). Three fields were randomly selected at a magnification of 200× magnification. Image Pro Plus software was used to calculate the mean optical density (MOD, MOD = integral optical density/measurement area).
Correlation Between PRDX1 Expression Level and Clinicopathological Characteristics
The chi-square test was performed to analyse the relationship between age, gender, pathological grade, tumor stage, T stage, N stage, and PRDX1 expression levels. Then, clinical features in the chi-square test (P < 0.05), and PRDX1 expression level were all included in the Cox proportional-hazards regression analysis. A nomogram based on factors with P < 0.05 in Cox regression analysis was generated to predict the prognosis of patients with OSCC, and the ROC curve and calibration chart were used to evaluate the prognostic performance of the nomogram.
Gene Set Enrichment Analysis (GSEA)
GSEA was performed using GSEA 4.1.0 software to analyse the gene ontology (GO), HALLMARK molecular mechanisms, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to PRDX1 in OSCC samples. The MSigDB gene set of ‘c5.all.v7.2.symbols.gmt’, ‘h.all.v7.2.symbols.gmt, ‘and ‘c2.cp.kegg.v7.2.symbols.gmt’ were used, and terms with P < 0.05 and FDR < 25% were considered to be significantly enriched.
Statistical Analysis
R studio (version 4.0.2), GraphPad Prism 8.0.2, and IBM SPSS® Statistics (version 25) software were used to analyse mRNA and protein expression data. The KM method, Log rank test, and Cox regression method were used to correlate clinicopathological data with the outcomes of patients with OSCC. Chi-square analysis was conducted to analyse the relationship between PRDX1 expression levels and clinical characteristics. The quantitative data in this study are expressed as mean ± standard deviation (SD). Statistical significance was set at P < 0.05. ***P < 0.001, **P < 0.01, *P < 0.05.
Results
The Expression Levels of PRDXs in TCGA Cohort
The expression of PRDXs between OSCC and normal tissues was compared in the TCGA cohort. The expression of PRDX1, PRDX4, and PRDX5 was significantly (P <0.05) higher in OSCC tissues, whereas that of PRDX2 was significantly higher in the ANTs. There was no significant difference in the expression of PRDX3 and PRDX6 between OSCC tissues and ANTs (Figure 1A–F). Then, the differences in PRDXs expression between non-metastatic and metastatic groups were analysed in the TCGA cohort. The results showed that except for PRDX5, the expression of PRDX1, PRDX2, PRDX3, PRDX4, and PRDX6 were significantly (P <0.05) higher in the metastatic group (Figure 2).
Prognostic Analysis Based on PRDX Expression
The effect of PRDXs on the overall survival rate of patients with OSCC was analysed in the TCGA cohort using the KM method. The results showed that patients with high expression of PRDX1, PRDX3, and PRDX6 had significantly lower overall survival rates (Figure 3A–F). Univariate Cox proportional-hazards regression analysis was further performed to evaluate the effect of PRDXs on the prognosis of patients with OSCC. PRDX1 and PRDX6 were found to affect the prognosis of patients as high-risk genes, and PRDX1 had the highest hazard ratio (hazard ratio: 1.950, P < 0.001) (Figure 3G).
PRDX1 Expression Level in Different Clinical Stages and Pathological Grades
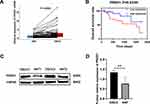
To further confirm the correlation between PRDX1 expression and prognosis in OSCC, qRT-PCR was used to analysed the mRNA expression of PRDX1 in the testing cohort consisting of 68 OSCC tissues and matched ANTs. The results showed that the expression level of PRDX1 mRNA in OSCC tissues was significantly upregulated (Figure 4A). The KM method revealed that upregulation of PRDX1 significantly affected the overall survival rate of these patients (Figure 4B). The result of Western blot also showed that compared to ANTs, PRDX1 protein level was increased in majority of the OSCC tissues (Figure 4C and D).
Furthermore, the differences in PRDX1 mRNA expression at different clinical stages and pathological grades were evaluated in the TCGA cohort. With the development of lymphatic metastasis, mRNA expression of PRDX1 was upregulated to varying degrees (Figure 5A), and it was also significantly upregulated in higher clinical stages and poorly differentiated samples (Figure 5B and C). qRT-PCR of the 68 OSCC tissues showed similar results (Figure 5D–F). In addition, as shown in Figure 6A and B, Western blot analysis revealed that protein expression of PRDX1 was upregulated in cases with severe lymphatic metastasis. IHC staining showed that high expression of PRDX1 was associated with poor differentiation (Figure 6C–F).
Relationship Between PRDX1 Expression Level and Clinicopathological Features
To evaluate the correlation between the expression level of PRDX1 and clinicopathological characteristics, chi-square test was performed on the clinical information and PRDX1 expression levels in TCGA cohort and confirmed the same in the testing cohort of 68 patients with OSCC. The results showed that gender, tumor stage, and N stage were significantly correlated with PRDX1 expression levels in TCGA cohort (Table 1). In the testing cohort, tumor stage, N stage and pathological grade were also found to be significantly correlated with PRDX1 expression level (Table 2). Next, the clinical features, including gender, pathological grade, tumor stage, and N stage, were all included into the Cox proportional-hazards regression analysis. The result showed that tumor stage and N stage were significantly associated with the prognosis of patients with OSCC (Figure 7A).
 |
Table 1 Correlation Between Expression Level of PRDX1and Clinical Characteristics in TCGA Cohort |
 |
Table 2 Correlation Between Expression Level of PRDX1and Clinical Characteristics in Testing Cohort |
Establishment of Nomogram
To further clarify the effect of PRDX1 on the prognosis of patients with OSCC, a nomogram was constructed with PRDX1 expression level, tumor stage and N stages to predict the 1-, 3-, and 5-year survival rates of patients with OSCC from the TCGA cohort, and was confirmed in the testing cohort (Figure 7B). The area under the curve (AUC) of ROC analysis in TCGA cohort was 0.620, 0.725, and 0.703 for 1-, 3-, and 5-year survival, respectively, and 0.678, 0.716, and 0.702 in the testing cohort (Figure 7C and D). This suggests that the nomogram optimally predicted the 1-, 3-, and 5-year survival rates of patients with OSCC in both TCGA and testing cohorts.
GSEA Between PRDX1 High and Low Expression Groups
GSEA was used to analyse the molecular mechanism of PRDX1 in OSCC. The results showed that high expression of PRDX1 not only improved cell respiration, oxidoreductase activity, electron transport chain, ATP synthesis, and other mitochondrial-related functions, but also significantly correlated with EMT, glycolysis, KRAS signaling, and ROS. Low expression of PRDX1 was significantly associated with immune-related functions, such as lymphocyte migration, CD4+ T cells, and tumor necrosis factor (Figure 8A and B).
 |
Figure 8 GSEA results showed the molecular mechanism of PRDX1 in OSCC. (A) GO; (B) Hallmarks; (C) KEGG pathways. |
In addition, downregulation of PRDX1 also participates in many immune-related signaling pathways, such as T and B cell receptor signaling pathways, as well as cell adhesion molecules and ECM receptor interaction. The upregulation of PRDX1 was significantly associated with basic transcription factors, spliceosomes, cell cycle, glycolysis, and DNA replication (Figure 8C).
Discussion
As the oral and maxillofacial region is abundant in lymphatic system, OSCC is very prone to lymphatic metastasis, and surgical methods have limited therapeutic effects on metastatic tumors.30,31 Therefore, it is very important to identify precise biomolecular targets for predicting metastasis, and treatment of OSCC.
Extensive research in the past has revealed the complex relationship between oxidative stress and tumorigenesis. Owing to the abnormal metabolism in tumors, tumor cells exhibit elevated ROS levels that is balanced by increasing antioxidant capacity, thereby promoting tumor development.32 Many studies have shown that PRDXs act as powerful antioxidant enzymes and organic hydroperoxide scavengers, as all PRDXs contain cysteine (Cys) as the primary site of oxidation and can be classified into 1-Cys and 2-Cys types based on the number of conserved Cys residues participating in the redox reaction.33 However, whether PRDXs can serve as prognostic biomarkers for OSCC remains unknown.
In this study, to explore the correlation between PRDXs and OSCC prognosis, gene expression data for 216 OSCC and 22 normal samples were obtained from TCGA data. We found that the expression of PRDX1, PRDX4, and PRDX5 was significantly upregulated in the OSCC samples compared to that in the normal samples, whereas the expression of PRDX2 was downregulated in OSCC samples, and there was no significant difference in the expression of PRDX6 between normal oral and OSCC samples. Our results for PRDX2 and PRDX6 were different from those of some previous studies.27,28,34 The functions of PRDX2 and PRDX6 in tumor progression remain controversial.35 PRDX2 and PRDX6 have been reported to be highly expressed in a variety of tumors including OSCC.28,29 High expression of PRDX2 or PRDX6 promotes metastasis of colorectal cancer.19,21 However, some studies have showed the down-regulation of PRDX2 can enhance the proliferation and migration of hepatocellular carcinoma, and is related to the poor prognosis of patients. Loss of PRDX6 in mice can enhance the susceptibility to skin tumorigenesis, whereas overexpression of PRDX6 in keratinocytes of transgenic mice has the opposite effect.36,37 Further research is needed to investigate the roles of PRDX2 and PRDX6 in OSCC. We further compared the differences in the expression of PRDXs between the OSCC metastatic and non-metastatic groups. The results showed that, except for PRDX5, the other five members of PRDX family were all upregulated in the metastatic group. KM method and univariate Cox proportional hazard regression analysis revealed that patients with high expression of PRDX1, PRDX3, and PRDX6 had significantly lower overall survival rates. Notably, PRDX1 was the only gene in the PRDX family that was not only highly expressed in OSCC tissues, and closely related to metastasis, but was also a high-risk factor affecting OSCC survival outcomes.
PRDX1 is an important member of the PRDX family, and its catalytic activity and protein sequence are different from those of the other antioxidants. PRDX1 has two conserved 2-Cys residues with redox activity, and the conserved N-terminal cysteine (Cys52-SH) is easily oxidized to Cys52-SOH by H2O2, while the H2O2 is reduced to water, reducing peroxides and other ROS molecules through thioredoxin, thus exerting a powerful role in scavenging free radicals.38 PRDX1 is highly expressed in many types of human malignant tumors, such as cervical cancer, hepatocellular carcinoma, and ovarian cancer, and affects the prognosis of patients.39–41 PRDX1 may also promote tumor invasion and metastasis in a variety of tumors, such as osteosarcoma and colorectal cancer.42,43 In addition, some studies have also revealed that PRDX1 is closely related to metastasis of OSCC.44,45
To further verify the correlation between PRDX1 and OSCC, we performed Western blot and qRT-PCR analyses in the testing cohort of 68 pairs of clinical OSCC tissues and ANTs. The results showed that PRDX1 was highly expressed in OSCC tissues and affected the overall survival rate of these patients, which was consistent with the results in the TCGA data. Moreover, we found that high expression of PRDX1 was also related to late tumor stage, severe lymph node metastasis, and poor differentiation in OSCC. Consistently, Chi-square analysis also showed that tumor stage, N stage, and pathological grade were significantly correlated with PRDX1 expression. This further demonstrates that PRDX1 may serve as a prognostic biomarker for OSCC.
In view of the prognostic value of PRDX1 in OSCC, we constructed a nomogram using PRDX1 levels in combination with high-risk clinicopathological factors that have a significant impact on the prognosis in TCGA cohort and confirmed the results in the testing. We found that pathological grade was significantly related to PRDX1 expression, but it was not a significant risk factor for patient survival. However, the ROC curve showed that the nomogram optimally predicted the 1-, 3-, and 5-year survival rates of patients with OSCC in both TCGA and testing cohorts.
To further explore the molecular mechanism of PRDX1 in OSCC, GSEA was performed. We observed that in addition to oxidoreductase activity, high expression of PRDX1 was mainly involved in mitochondrial-related functions, such as ATP biosynthetic process, cellular respiration, and mitochondrial protein complex. Some studies have shown that mitochondria play an important role in the development of tumors, although the mitochondrial genome in most tumors changes and causes mitochondrial dysfunction. Increasing evidence suggests that abnormal mitochondria in cancer cells are involved in mitochondrial apoptosis, adaptation to the hypoxic microenvironment, glycolysis or oxidative phosphorylation, and metastasis.46,47 In addition, the results of GSEA also showed that PRDX1 is related to many pathways and hallmarks related to cancer, such as cell cycle, DNA replication, E2F targets, and EMT. As a transcription factor, E2F plays an important role in the occurrence and development of cancer by regulating the cell cycle and apoptosis.48 PRDX1 promotes the EMT process of gastric cancer by inhibiting the expression of E-cadherin, and promotes cell proliferation and metastasis by enhancing Akt/mTOR in human osteosarcoma cells.49,50 Our previous study also confirmed that PRDX1 promotes the invasion and migration of tobacco-related OSCC by regulating the EMT process.51 PRDX1 knockdown significantly inhibited cervical metastasis rate of tongue cancer, upregulated E-cadherin expression, and downregulated the expression of vimentin and Snail in the tongue and lymph nodes of mice.52 However, low expression of PRDX1 is mainly associated with lymphocyte migration, immune receptor activity, B-cell and T-cell receptor signaling pathways, inflammatory responses, and other immune-related functions or pathways. These results further confirm that high expression of PRDX1 promotes OSCC.
Conclusion
In summary, PRDX1 is significantly related to tumor stage, lymph node metastasis, and pathological grade, and affects the prognosis of patients with OSCC. The nomogram constructed based on PRDX1 expression showed good performance in predicting the prognosis of patients with OSCC. Our findings have clear implications for the application of PRDX1 as a biomarker for predicting lymph node metastasis and prognosis of patients with OSCC. Further clinical studies are required to validate these conclusions.
Abbreviations
HNC, head and neck cancer; OSCC, oral squamous cell carcinoma; ROS, reactive oxygen species; PRDXs, peroxiredoxins; Trx, thioredoxin; TCGA, The Cancer Genome Atlas; ANTs, adjacent noncancerous tissues; TPM, transcripts per million; KM, Kaplan-Meier; RIPA, radioimmunoprecipitation assay; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; PVDF, polyvinylidene difluoride; qRT, quantitative reverse transcription; IHC, Immunohistochemical; HRP, horseradish peroxidase; MOD, mean optical density; GSEA, gene set enrichment analysis; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; AUC, area under the curve; Cys, cysteine.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethical Approval Statement
This research complied with the Declaration of Helsinki and was approved by the Research Ethics Committee of the Beijing Stomatological Hospital of Capital Medical University (Approval No. CMUSH-IRB-KJ-PJ-2018-01). All patients agreed to use their samples in this study and signed written informed consent.
Acknowledgments
This study was supported by the Beijing Municipal Natural Science Foundation of China (Grant No: 7192075).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that there is no conflict of interest that can affect the publication of this article.
References
1. Chow LQM, Longo DL. Head and Neck Cancer. N Engl J Med. 2020;382(1):60–72. doi:10.1056/NEJMra1715715
2. Sarode GS, Sarode SC, Maniyar N, Anand R, Patil S. Oral cancer databases: a comprehensive review. J Oral Pathol Med. 2018;47(6):547–556. doi:10.1111/jop.12667
3. Sharma A, Kim JW, Paeng JY. Clinical analysis of neck node metastasis in oral cavity cancer. J Korean Assoc Oral Maxillofac Surg. 2018;44(6):282–288. doi:10.5125/jkaoms.2018.44.6.282
4. Bugshan A, Farooq I. Oral squamous cell carcinoma: metastasis, potentially associated malignant disorders, etiology and recent advancements in diagnosis. F1000Res. 2020;9:229. doi:10.12688/f1000research.22941.1
5. Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5(1):28. doi:10.1038/s41392-020-0134-x
6. Garza-Lombo C, Pappa A, Panayiotidis MI, Franco R. Redox homeostasis, oxidative stress and mitophagy. Mitochondrion. 2020;51:105–117. doi:10.1016/j.mito.2020.01.002
7. Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168(4):644–656. doi:10.1016/j.cell.2017.01.002
8. Galadari S, Rahman A, Pallichankandy S, Thayyullathil F. Reactive oxygen species and cancer paradox: to promote or to suppress? Free Radic Biol Med. 2017;104:144–164. doi:10.1016/j.freeradbiomed.2017.01.004
9. Panieri E, Santoro MM. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death Dis. 2016;7(6):e2253. doi:10.1038/cddis.2016.105
10. Sullivan LB, Chandel NS. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014;2:17. doi:10.1186/2049-3002-2-17
11. Marengo B, Nitti M, Furfaro AL, et al. Redox homeostasis and cellular antioxidant systems: crucial players in cancer growth and therapy. Oxid Med Cell Longev. 2016;2016:6235641. doi:10.1155/2016/6235641
12. Kim Y, Jang HH. The role of peroxiredoxin family in cancer signaling. J Cancer Prev. 2019;24(2):65–71. doi:10.15430/JCP.2019.24.2.65
13. Rhee SG, Woo HA, Kang D. The Role of peroxiredoxins in the transduction of H2O2 signals. Antioxid Redox Signal. 2018;28(7):537–557. doi:10.1089/ars.2017.7167
14. Hampton MB, Vick KA, Skoko JJ, Neumann CA. Peroxiredoxin involvement in the initiation and progression of human cancer. Antioxid Redox Signal. 2018;28(7):591–608. doi:10.1089/ars.2017.7422
15. Ismail T, Kim Y, Lee H, Lee DS, Lee HS. Interplay between mitochondrial peroxiredoxins and ROS in cancer development and progression. Int J Mol Sci. 2019;20(18). doi:10.3390/ijms20184407
16. Song IS, Kim HK, Jeong SH, et al. Mitochondrial peroxiredoxin III is a potential target for cancer therapy. Int J Mol Sci. 2011;12(10):7163–7185. doi:10.3390/ijms12107163
17. Whitaker HC, Patel D, Howat WJ, et al. Peroxiredoxin-3 is overexpressed in prostate cancer and promotes cancer cell survival by protecting cells from oxidative stress. Br J Cancer. 2013;109(4):983–993. doi:10.1038/bjc.2013.396
18. Liu Y, Kwon T, Kim JS, et al. Peroxiredoxin V reduces beta-Lapachone-induced apoptosis of colon cancer cells. Anticancer Res. 2019;39(7):3677–3686. doi:10.21873/anticanres.13516
19. Peng L, Wang R, Shang J, Xiong Y, Fu Z. Peroxiredoxin 2 is associated with colorectal cancer progression and poor survival of patients. Oncotarget. 2017;8(9):15057–15070. doi:10.18632/oncotarget.14801
20. Ahn HM, Yoo JW, Lee S, et al. Peroxiredoxin 5 promotes the epithelial-mesenchymal transition in colon cancer. Biochem Biophys Res Commun. 2017;487(3):580–586. doi:10.1016/j.bbrc.2017.04.094
21. Huang WS, Huang CY, Hsieh MC, et al. Expression of PRDX6 correlates with migration and invasiveness of colorectal cancer cells. Cell Physiol Biochem. 2018;51(6):2616–2630. doi:10.1159/000495934
22. Ramasamy P, Larkin, AM, Linge A, et al. PRDX3 is associated with metastasis and poor survival in uveal melanoma. J Clin Pathol. 2020;73(7):408–412. doi:10.1136/jclinpath-2019-206173
23. Wang G, Zhong WC, Bi YH, et al. The prognosis of peroxiredoxin family in breast cancer. Cancer Manag Res. 2019;11:9685–9699. doi:10.2147/CMAR.S229389
24. Xu R, Pan J, Mei J, Zhang Q. Systematic characterization of prognostic values of peroxiredoxin family in gastric cancer. Biomed Res Int. 2020;2020:3948183. doi:10.1155/2020/3948183
25. Sienko J, Teliga-Czajkowska J, Przytula E, Czajkowski K, Smolarczyk R, Nowis D. Peroxiredoxin-1 as a prognostic factor in patients with ovarian cancer. Ann Agric Environ Med. 2019;26(3):415–419. doi:10.26444/aaem/105899
26. Lopez-Grueso MJ, Lagal DJ, Garcia-Jimenez AF, et al. Knockout of PRDX6 induces mitochondrial dysfunction and cell cycle arrest at G2/M in HepG2 hepatocarcinoma cells. Redox Biol. 2020;37:101737. doi:10.1016/j.redox.2020.101737
27. Lee EY, Kang JY, Kim KW. Expression of cyclooxygenase-2, peroxiredoxin I, peroxiredoxin 6 and nuclear factor-kappaB in oral squamous cell carcinoma. Oncol Lett. 2015;10(5):3129–3136. doi:10.3892/ol.2015.3705
28. Chuerduangphui J, Ekalaksananan T, Heawchaiyaphum C, Vatanasapt P, Pientong C. Peroxiredoxin 2 is highly expressed in human oral squamous cell carcinoma cells and is upregulated by human papillomavirus oncoproteins and arecoline, promoting proliferation. PLoS One. 2020;15(12):e0242465. doi:10.1371/journal.pone.0242465
29. Huang CF, Sun ZJ, Zhao YF, Chen XM, Jia J, Zhang WF. Increased expression of peroxiredoxin 6 and cyclophilin A in squamous cell carcinoma of the tongue. Oral Dis. 2011;17(3):328–334. doi:10.1111/j.1601-0825.2010.01730.x
30. Irani S. Distant metastasis from oral cancer: a review and molecular biologic aspects. J Int Soc Prev Community Dent. 2016;6(4):265–271. doi:10.4103/2231-0762.186805
31. Zanoni DK, Montero PH, Migliacci JC, et al. Survival outcomes after treatment of cancer of the oral cavity (1985-2015). Oral Oncol. 2019;90:115–121. doi:10.1016/j.oraloncology.2019.02.001
32. Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12(12):931–947. doi:10.1038/nrd4002
33. Rhee SG. Overview on peroxiredoxin. Mol Cells. 2016;39(1):1–5. doi:10.14348/molcells.2016.2368
34. Heawchaiyaphum C, Pientong C, Phusingha P, et al. Peroxiredoxin-2 and zinc-alpha-2-glycoprotein as potentially combined novel salivary biomarkers for early detection of oral squamous cell carcinoma using proteomic approaches. J Proteomics. 2018;173:52–61. doi:10.1016/j.jprot.2017.11.022
35. Nicolussi A, D’Inzeo S, Capalbo C, et al. The role of peroxiredoxins in cancer. Mol Clin Oncol. 2017;6(2):139–153. doi:10.3892/mco.2017.1129
36. Bai B, Lin Y, Hu J, et al. Peroxiredoxin2 downregulation enhances hepatocellular carcinoma proliferation and migration, and is associated with unfavorable prognosis in patients. Oncol Rep. 2019;41(3):1539–1548. doi:10.3892/or.2019.6977
37. Rolfs F, Huber M, Gruber F, et al. Dual role of the antioxidant enzyme peroxiredoxin 6 in skin carcinogenesis. Cancer Res. 2013;73(11):3460–3469. doi:10.1158/0008-5472.CAN-12-4369
38. Sun CC, Dong WR, Shao T, et al. Peroxiredoxin 1 (Prx1) is a dual-function enzyme by possessing Cys-independent catalase-like activity. Biochem J. 2017;474(8):1373–1394. doi:10.1042/BCJ20160851
39. Lu E, Hu X, Pan C, Chen J, Xu Y, Zhu X. Up-regulation of peroxiredoxin-1 promotes cell proliferation and metastasis and inhibits apoptosis in cervical cancer. J Cancer. 2020;11(5):1170–1181. doi:10.7150/jca.37147
40. Sun QK, Zhu JY, Wang W, et al. Diagnostic and prognostic significance of peroxiredoxin 1 expression in human hepatocellular carcinoma. Med Oncol. 2014;31(1):786. doi:10.1007/s12032-013-0786-2
41. Zheng MJ, Wang J, Wang HM, et al. Decreased expression of peroxiredoxin1 inhibits proliferation, invasion, and metastasis of ovarian cancer cell. Onco Targets Ther. 2018;11:7745–7761. doi:10.2147/OTT.S175009
42. Wang Y, Liu M, Yang P, Peng H. Peroxiredoxin 1 (PRDX1) suppresses progressions and metastasis of osteosarcoma and fibrosarcoma of bone. Med Sci Monit. 2018;24:4113–4120. doi:10.12659/MSM.908736
43. Li HX, Sun XY, Yang SM, Wang Q, Wang ZY. Peroxiredoxin 1 promoted tumor metastasis and angiogenesis in colorectal cancer. Pathol Res Pract. 2018;214(5):655–660. doi:10.1016/j.prp.2018.03.026
44. Jiang Y, Cao W, Wu K, et al. LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus. J Exp Clin Cancer Res. 2019;38(1):365. doi:10.1186/s13046-019-1364-z
45. Zhang N, Zeng L, Wang S, et al. LncRNA FER1L4 promotes oral squamous cell carcinoma progression via targeting miR-133a-5p/Prx1 axis. Onco Targets Ther. 2021;14:795–806. doi:10.2147/OTT.S277351
46. Onishi M, Yamano K, Sato M, Matsuda N, Okamoto K. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021;40(3):e104705. doi:10.15252/embj.2020104705
47. Guerra F, Arbini AA, Moro L. Mitochondria and cancer chemoresistance. Biochim Biophys Acta Bioenerg. 2017;1858(8):686–699. doi:10.1016/j.bbabio.2017.01.012
48. Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer. 2019;19(6):326–338. doi:10.1038/s41568-019-0143-7
49. Yu W, Wu J, Ning ZL, Liu QY, Quan RL. High Expression of peroxiredoxin 1 is associated with epithelial-mesenchymal transition marker and poor prognosis in gastric cancer. Med Sci Monit. 2018;24:2259–2270. doi:10.12659/MSM.908722
50. Cai AL, Zeng W, Cai WL, et al. Peroxiredoxin-1 promotes cell proliferation and metastasis through enhancing Akt/mTOR in human osteosarcoma cells. Oncotarget. 2018;9(9):8290–8302. doi:10.18632/oncotarget.23662
51. Niu W, Zhang M, Chen H, et al. Peroxiredoxin 1 promotes invasion and migration by regulating epithelial-to-mesenchymal transition during oral carcinogenesis. Oncotarget. 2016;7(30):47042–47051. doi:10.18632/oncotarget.9705
52. Wang M, Niu W, Qi M, et al. Nicotine promotes cervical metastasis of oral cancer by regulating peroxiredoxin 1 and epithelial-mesenchymal transition in mice. Onco Targets Ther. 2019;12:3327–3338. doi:10.2147/OTT.S194129
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.