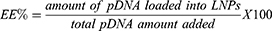Back to Journals » International Journal of Nanomedicine » Volume 17
Ionizable Lipid Nanoparticle-Mediated Delivery of Plasmid DNA in Cardiomyocytes
Authors Scalzo S, Santos AK , Ferreira HAS , Costa PA, Prazeres PHDM , da Silva NJA , Guimarães LC, e Silva MDM, Rodrigues Alves MTR, Viana CTR, Jesus ICG, Rodrigues AP , Birbrair A, Lobo AO , Frezard F, Mitchell MJ, Guatimosim S, Guimaraes PPG
Received 18 March 2022
Accepted for publication 19 June 2022
Published 30 June 2022 Volume 2022:17 Pages 2865—2881
DOI https://doi.org/10.2147/IJN.S366962
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yan Shen
Sérgio Scalzo,1 Anderson K Santos,1 Heloísa AS Ferreira,1 Pedro A Costa,1 Pedro HDM Prazeres,2 Natália JA da Silva,1 Lays C Guimarães,1 Mário de Morais e Silva,1 Marco TR Rodrigues Alves,1 Celso TR Viana,1 Itamar CG Jesus,1 Alice P Rodrigues,1 Alexander Birbrair,2 Anderson O Lobo,3 Frederic Frezard,1 Michael J Mitchell,4 Silvia Guatimosim,1 Pedro Pires Goulart Guimaraes1
1Department of Physiology and Biophysics, Federal University of Minas Gerais, Belo Horizonte, MG, Brazil; 2Department of General Pathology, Federal University of Minas Gerais, Belo Horizonte, MG, Brazil; 3Department of Materials Engineering, Federal University of Piauí, Teresina, PI, Brazil; 4Department of Bioengineering, University of Pennsylvania, Philadelphia, PA, USA
Correspondence: Pedro Pires Goulart Guimaraes, Department of Physiology and Biophysics, Federal University of Minas Gerais, Belo Horizonte, MG, Brazil, Tel +55 031 3409-2948, Email [email protected]
Introduction: Gene therapy is a promising approach to be applied in cardiac regeneration after myocardial infarction and gene correction for inherited cardiomyopathies. However, cardiomyocytes are crucial cell types that are considered hard-to-transfect. The entrapment of nucleic acids in non-viral vectors, such as lipid nanoparticles (LNPs), is an attractive approach for safe and effective delivery.
Methods: Here, a mini-library of engineered LNPs was developed for pDNA delivery in cardiomyocytes. LNPs were characterized and screened for pDNA delivery in cardiomyocytes and identified a lead LNP formulation with enhanced transfection efficiency.
Results: By varying lipid molar ratios, the LNP formulation was optimized to deliver pDNA in cardiomyocytes with enhanced gene expression in vitro and in vivo, with negligible toxicity. In vitro, our lead LNP was able to reach a gene expression greater than 80%. The in vivo treatment with lead LNPs induced a twofold increase in GFP expression in heart tissue compared to control. In addition, levels of circulating myeloid cells and inflammatory cytokines remained without significant changes in the heart after LNP treatment. It was also demonstrated that cardiac cell function was not affected after LNP treatment.
Conclusion: Collectively, our results highlight the potential of LNPs as an efficient delivery vector for pDNA to cardiomyocytes. This study suggests that LNPs hold promise to improve gene therapy for treatment of cardiovascular disease.
Keywords: lipid nanoparticles, ionizable lipids, pDNA delivery, heart, cardiomyocytes
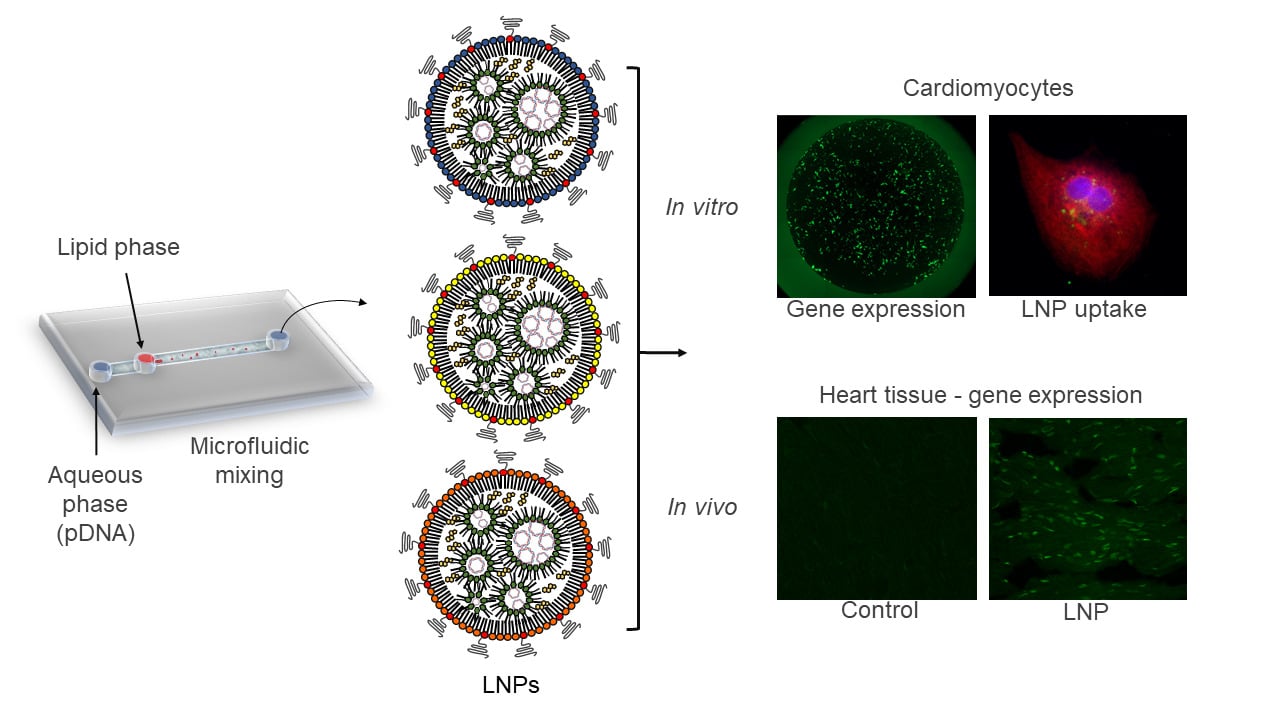
Graphical Abstract:
Introduction
Cardiovascular diseases (CVD) refer to a group of disorders of blood vessels and the heart.1–3 Despite all the progress and clinical innovations made over the past decades, CVD still have a high prevalence and represent the leading cause of death worldwide, with a significant impact on the healthcare system costs of many countries.4,5 With an estimated 17.9 million deaths per year, CVD are considered part of chronic non-communicable diseases, representing approximately 32% of total deaths worldwide in 2019.3 To address this challenge and to overcome the limitations of conventional therapies, much attention has been given to advances in delivery systems, which are associated with the knowledge of the target’s signaling pathways and biological barriers, and are being exploited to develop more effective gene therapies.4 The potential of gene therapy has provided unprecedented opportunities to explore challenges from early treatments.4,6 It is also a promising approach to be applied in cardiac regeneration after myocardial infarction and gene modulation for inherited.7–11 While several preclinical and clinical studies have shown the efficacy of gene therapy in many tissues and cells, the therapeutic efficacy of this approach remains a challenge in the heart tissue.8 This is likely due to several factors, including nucleic acid degradation, inactivation by serum proteins, loss of therapeutic effect over time, and insufficient delivery to the cardiac tissue.12 Moreover, the cardiac sarcolemma represents an additional barrier to nucleic acid delivery.8 Thus, one of the greatest challenges faced is the optimal gene delivery vector, which should be safe, effective, and display selectivity for the cardiac tissue.11
Both non-viral and recombinant viral delivery methods for gene therapy have now been established for their use in clinical trials.11 Although viral vectors such as lentiviruses and adenovirus have been used for nucleic acid delivery, their use is limited due to immune responses raised against viral proteins.13 Thus, novel strategies are needed to improve gene delivery properties and transfection of gene therapy in the heart, and the use of nanoparticles (NPs) as carriers have proven a promising approach to be used in CVD models.14,15 The entrapment of nucleic acids in non-viral vectors, such as NPs, represents an attractive approach for safe and effective delivery and transfection efficiency, which can provide stability, longer circulation, avoid renal clearance from the blood due to size-based effects, and prevent nonspecific interactions via PEGylation of NPs.16 Additionally, NPs can also extravasate from the bloodstream to reach target tissues via various targeting-based approaches, mediate cell entry, and provide endosomal escape.17
In this context, ionizable lipids have been successfully used to formulate NPs for mRNA, and DNA delivery.18 Amine-containing ionizable lipids have been designed to enhance transfection and reduce toxicity compared to cationic lipids.19 Ionizable lipids positively charged at low pH enables the complexation with negatively charged nucleic acid in nanoparticle formulation in acidic buffers19 Furthermore, ionizable lipids are neutral at physiological pH and positively charged into endosomes, exhibiting special mechanisms such as the “proton-sponge effect” and endosome-lytic properties to endosomal escape.20,21 Indeed, a high degree of protonation facilitates the endosomal escape of RNA, DNA, and the traffic through the cytoplasm to the nucleus for gene expression.20,21 In addition to an ionizable lipid, other excipients are also commonly used to formulate lipid nanoparticles (LNPs), such as a) a phospholipid to support the LNP bilayer structure and aid in endosomal escape,22 b) cholesterol to improve the stability of the LNP bilayer and induce membrane fusion,23 and c) a lipid-polyethylene glycol (PEG) conjugate to reduce LNP aggregation and nonspecific endocytosis by immune cells.24
It was hypothesized that LNPs can deliver not only mRNA but also plasmid DNA (pDNA) safely and effectively, especially to hard-to-transfect cardiomyocytes. Thus, herein, our objective was to develop and screen a mini-library of pDNA-loaded LNPs for enhanced transfection efficiency in cardiomyocytes. By varying lipid ratios, we identified an optimized LNP formulation that significantly enhanced gene expression in cardiomyocytes in vitro and in vivo, with negligible toxicity.
Materials and Methods
Plasmid DNA (pDNA)
E. coli Stbl3 transformed with pZsGreen-N1 (Clontech laboratories) that encodes ZsGreen, a green fluorescent protein (GFP), were grown in LB medium and the plasmid was purified by PureLink™ HiPure Plasmid Maxiprep Kit (Invitrogen), following the manufacturer’s instructions and quantified by NanoDrop-OneC-UV-Vis (Thermo Fisher).
Lipid Nanoparticle (LNP) Formulation
To formulate LNPs, the ethanolic phase containing ionizable lipids and excipients was mixed with an aqueous phase of pDNA using a microfluidic device as previously described.18 Ionizable lipid C12-200 (synthesized as previously described25), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE, Avanti Polar Lipids, no. 850725P), cholesterol (Avanti Polar Lipids, no. 700100P) and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethyleneglycol)-2000] (ammonium salt) (C14-PEG 2000, Avanti Polar Lipids, no. 880150P) were prepared in ethanol (10 to 100 mg/mL) and mixed at predetermined molar ratios based on ionizable lipid:pDNA ratio, which was varied in LNP formulations (10:1 or 20:1) as shown in Table 1. The aqueous phase containing plasmid DNA coding for green fluorescent protein (GFP) was prepared in 10 mM citrate in a volume ratio of citrate phase:ethanolic phase of 2.5:1. Citrate buffer (pH 3.0, Teknova) was used to ensure the ionization of C12-200 and interaction with negatively charged pDNA during mixing. Both the inlets were connected to programmable syringe pumps (Harvard Apparatus) and the solutions were injected into the microfluidic devices at a 2.5:1 ratio (citrate phase:ethanolic phase). LNPs were collected and then dialyzed against 2L PBS in a 20,000 MWCO dialysis cassette for 2h. Afterward, the LNPs were filtered through a 0.22-µm filter to be assessed in vitro and in vivo. To prepare fluorescent LNPs, regular cholesterol was replaced by fluorescent cholesterol (23-(dipyrrometheneboron difluoride)-24-norcholesterol (ex/em 495/507 nm), Avanti Polar Lipids no. 810255P), following the same protocol.
 |
Table 1 Characterization of pDNA Encapsulated LNPs with Different C12-200:pDNA Ratio and Composition |
Characterization of LNP
LNP hydrodynamic diameter (HD), polydispersity index (PDI), and zeta potential (ZP) were measured using a Zetasizer Nano ZS machine (Malvern Instrument). The structure analysis of LNPs was performed via cryogenic-transmission electron microscopy (Cryo-TEM). Vitrified samples were examined using Tecnai G2-20 - FEI SuperTwin 200 kV at the Center for Acquisition and Processing of Images (Centro de Aquisição e Processamento de Imagens – UFMG). Further analysis using NanoDrop and Qubit dsDNA HS Assay Kit was performed to quantify dsDNA and to determine the encapsulation efficiency (EE) of each LNP. pDNA EE was calculated from the following equation:
Animal Models
All procedures and experiments were performed following the guidelines and policies of the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. Neonatal (P1-3) Wistar rats were provided by the Animal Facility of the Institute of Biological Sciences at the Federal University of Minas Gerais (ICB-UFMG) and C57BL/6 mice were provided by Biotério Central, Federal University of Minas Gerais. The treatment protocol was previously approved by the Institutional Animal Care and Use Committee at UFMG (IACUC-UFMG 138/2018 and 177/2020).
Neonatal Ventricular Myocyte Isolation
Cardiac tissue was removed from 1-3 days-old Wistar rats, as previously described.26 After 24h, cells were rinsed with 200 µL/well of M199 medium and fed for 48h with a regular culture medium, now including 20 µg/mL cytosine-D-arabinofuranoside to inhibit the growth of noncardiomyocytes. Neonatal cardiomyocytes were used on the fourth day of cultivation.
Cardiomyocyte Transfection
To screen LNPs, neonatal cardiomyocytes were plated in a 96-well plate (1 × 104 cells/well) using M199 medium with 10% FBS and incubated with different LNPs’ formulations (Table 1) at 0.4–0.8 µg of pDNA. To assess the transfection efficiency of lead LNP, cardiomyocytes were plated in a 96-well plate (1 × 104 cells/well) using M199 medium with 10% FBS and incubated with different LNPs formulations (Table 1) at varying amounts of pDNA (0.00625–0.8 µg). All image fluorescence was carried out at the Center for Acquisition and Processing of Images (CAPI-UFMG) using Cytation 5 Cell Imaging (Biotek) with 4× or 40× objective and a filter-based 633 nm, 488 nm, and 358 nm channel. For GFP quantification, the fluorescence intensity images were selected as a green channel and converted to an 8-bit grayscale image, and normalized by the non-transfected cells images. Otsu’s method was applied to calculate the optimal threshold pixel intensity.27 The mean pixel intensity was measured and plotted.
LNP Uptake by Cardiomyocytes
To assess LNP uptake, cardiomyocytes were incubated with LNP4-6 (0.2 μg of pDNA) formulated with fluorescent cholesterol (ex/em 488/520 nm) and fixed with 100 µL/well of 4% paraformaldehyde (PFA) in a 96-well plate at 2h, 4h, and 6h time points. Cells were stained with 50 µL of anti-alpha-actinin antibody (1:150, Sigma, no. A7811) with goat anti-mouse Alexa Fluor 633 (ex/em 633/650 nm, 1:400, Thermo Fisher Scientific, no. A11001) as the secondary antibody, and the nuclei were stained with 50 µL of DAPI (ex/em 340/488 nm, 1:500, stock solution:5mg/mL, Sigma, no. D8417). Images were acquired at the CAPI-UFMG using Cytation 5 Cell Imaging (Biotek) with a 40× objective.
In situ Determination of pKa Using TNS
To determine the pKa of LNPs, a fluorescent reagent 6-(p-Toluidino)-2-naphthalenesulfonic acid sodium salt (TNS) (Sigma-Aldrich no. T9792) was used as previously described.28 Briefly, stock buffered solutions of 150 mM sodium chloride, 20 mM sodium phosphate, 25 mM sodium citrate, and 20 mM sodium acetate were prepared and adjusted to reach pH values ranging from 2 to 12 in increments of 0.5. Then, 5 µL of pDNA-LNP (~290 ng/µL) was added in 140 µL of each pH-adjusted solution in a 96-well plate in triplicate. Following, 5 µL of 0.18 mM stock solution of TNS, prepared in Milli-q water, was then added to each well to reach a final TNS concentration of 6 µM. The fluorescence of the samples (ex/em 325/435 nm) was then measured on a plate reader VarioskanTM Flash (Thermo Scientific). The pKa values were calculated by applying the Henderson–Hasselbalch equation using a sigmoidal curve-fit analysis to establish 50% of maximal fluorescence intensity, which reflects 50% of protonation.
Cell Viability by Resazurin Reduction
To investigate the cell toxicity after treatment with LNPs formulations, the resazurin reduction method was used, as previously described.29 Cardiomyocytes were treated with 10 µL of LNP formulations (for a final amount of 0.2 µg of pDNA in each well) for 48h, followed by the addition of 10 µL of Resazurin (5 µg/mL, Sigma, no. R7017) for 4h. Fluorescence was measured using the VarioskanTM Flash reader (ex/em 530/590 nm).
In vivo Transfection Efficiency
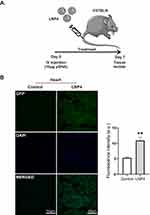
To assess delivery and transfection efficiency in vivo, 8- to 10-week-old female C57BL/6 mice were used (n=4). For this, mice were treated with PBS or pDNA coding for GFP encapsulated in LNP4 (10 µg total pDNA) via tail vein injection. After 7 days, mice were euthanized and the heart was harvested to assess the transfection efficiency and toxicity of LNPs. Tissues were immersed in cryomolds containing OCT – optimal cutting temperature (Tissue Tek) and frozen at −80 °C for subsequent fluorescence analysis. Tissues were processed using a cryostat at a thickness of 8mm. Images from the sections were acquired using the Zeiss LSM 880 Meta confocal microscope with an inverted objective at 60×/1.3 and a filter-based 488 nm channel, located in the CAPI-UFMG.
Western Blotting
To investigate the effect of the treatment with LNP4 in the expression of proteins involved in cardiomyocyte function (SERCA2) and survival (AKT and ERK1/2), adult ventricular myocytes were harvested in ice-cold lysis buffer (in mmol/L: 100 NaCl, 50 Tris-base, 5 EDTA,·2 H2O, 50 Na4P2O7, 10 H2O, and 1 MgCl2, pH 8.0) containing 0.3% Triton X-100, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 20 mM NaF, enriched with 10 µL of protease (1% (v/v), no. P8340; Sigma-Aldrich) and 10 µL of phosphatase inhibitors cocktail (1% (v/v), no. P0044; SigmaAldrich). Then, 30 µg of protein were separated by SDS-PAGE followed by Western blotting. Immunodetection was carried out using enhanced chemiluminescence detected with LAS 4000 equipment (GE Healthcare Life Science). Protein levels were expressed as a ratio of optical densities. GAPDH, AKT, or ERK1/2 total expression was used as a control for any variations in protein loading.
qRT-PCR
After in vivo treatment with LNP4, gene expression of inflammatory cytokines was analyzed via qRT-PCR. RNA was isolated from the heart using Trizol reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions, and quantified spectrophotometrically using the Nanodrop Lite (Thermo Fisher Scientific). Afterward, 1 µg of RNA was used for first-strand cDNA synthesis [Reverse Transcriptase (RT)] using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qRT-PCR was performed with diluted cDNA using PowerUp SYBR Green Master Mix (Applied Biosystems) on QuantStudio 3 (Applied Biosystems). Sequence primers were IL-6 forward 5’ GGTGCCCTGCCAGTATTCTC 3’, IL-6 reverse 5’ GGCTCCCAACACAGGATGA 3’; TNFα forward 5’ CCCTCACACTCAGATCATCTTCT 3’, TNFα reverse 5’ GCTACGACGTGGGCTACAG 3’, actin forward 5’ GGC TGTATTCCCCTCCATCGB 3’, actin reverse 5’ CCAGTTGGTAACAATGCCATGT 3’. Ct values were recorded for each gene and the results were normalized to actin.30
Flow Cytometry Studies
To characterize myeloid cell types in the heart after treatment with LNP4 formulation or control according to the protocol described earlier, single-cell suspensions from saline perfused hearts were minced and digested in DMEM with Collagenase I at 3 mg/mL for 45 minutes at 37°C. To deactivate the enzyme, the same amount of fetal bovine serum was added to each sample. Samples were filtered through 40 μm cell strainers. Red blood cell lysis was performed with ACK lysis buffer (Thermo Fisher Scientific). Samples were washed once with 200 µL of FACS buffer (PBS with 2% FBS and 2 mM EDTA) and resuspended in 100 μL of the same solution, and then incubated with a mix of 30 μL/well of antibodies CD45-PerCP-Cy5.5 (1:200, BioLegend, no. 109828), CD11b-SB600 (1:500, Invitrogen, no. 63–0112-82), F4/80-APC (1:200, Invitrogen, no. 17–4801-82), Gr1-Biotin (1:200, Biolegend, no. 108404), and Streptavidin-PacificOrange (1:500, Invitrogen no. S32365) at RT for 20 minutes in the dark . The cells were then analyzed using a BD LSRII flow cytometer (BD Biosciences). Neutrophils were gated as CD11b+Ly6Ghigh. Monocytes were gated as CD11b+Ly6ChighMHCIIlow. Macrophages were gated as CD11b+Ly6G−Ly6Clow F4/80+ cells.
Quantification and Statistical Analysis
Data are presented as mean ± SEM of at least three independent experiments. For statistical comparison, Student t-test, and one-way ANOVA were used followed by Dunnett’s Multiple Comparison post hoc test. The level of significance was set to values of p<0.05.
Results
Synthesis, Characterization, and Optimization of LNPs
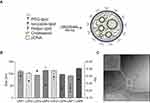
Distinct LNPs were formulated using mixtures of an ionizable lipid (C12-200), dioleoylphosphatidylethanolamine (DOPE), cholesterol (CHOL), PEG-lipid conjugate, C14-PEG2000 at different lipid ratios (Table 1). LNPs were formed in the presence of pDNA via microfluidic mixing (Figure 1A).18
Here, it was hypothesized that LNPs made from these components could be optimized for pDNA delivery in cardiomyocytes. Therefore, an LNP mini-library complexed with pDNA coding for GFP was synthesized and optimized, by varying C12-200:pDNA ratio, DOPE, and CHOL ratios (Table 1).
LNPs were characterized by size, pKa, and pDNA encapsulation efficiency. The hydrodynamic diameter of all LNPs was between 91.8 nm and 122.6 nm, while their polydispersity index and encapsulation efficiency ranged from 0.216 to 0.337 and 71% to 94%, respectively (Figure 1B and Table 1). Six of the eight LNP formulations possessed over 80% of encapsulation efficiency. Cryo-TEM micrograph showed that LNPs formed monodisperse LNPs, in agreement with DLS findings, with a dense core and spherical morphology (Figure 1C). Negative surface charges between −3.2 mV and −9.2 mV were observed for LNPs with a 10:1 ratio (C12-200:pDNA) while the LNPs with a 20:1 ratio (LPN5-8) possessed positive charge values (between 10.8 mV and 16.3 mV) (Table 1).
In vitro Cardiomyocytes Transfection by LNPs
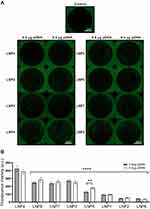
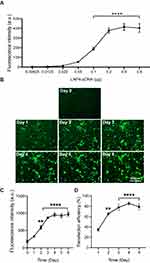
To determine the ability of LNPs to transfect cardiomyocytes and evaluate the formulation that enables more efficient pDNA delivery to the cardiomyocytes, eight unique LNP formulations were developed. To assess transfection efficiency, LNPs were complexed with pDNA coding for GFP. Thus, cardiomyocytes were treated with LNPs at doses of 0.4 and 0.8 µg of pDNA. After 48h, GFP expression was assessed by fluorescence acquisition using Cytation 5 Cell Imaging. All LNP formulations were able to transfect myocyte cells (Figure 2A and B), yet with variable fluorescence intensity (Figure 2B), which reflects both LNP delivery and pDNA functionality. As shown in Figure 2B, LNP4 exhibited enhanced GFP fluorescence compared to other LNP formulations achieving around 1.3- and 10-fold increased fluorescence than the second-best formulation candidate (LNP8), and the lowest-performing formulation (LNP6), respectively (Figure 2B). Of note, LNPs with higher DOPE molar ratio (Figure 3A and B) and lower cholesterol molar ratio (Figure 3C and D) induced enhanced GFP expression, independently of the amount of pDNA used (Figures 2B and 3). Collectively, these results indicate LNP4 as the top-performing formulation.
LNPs Cellular Uptake, pKa Measurement, and Cell Viability Assay
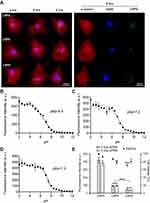
To confirm the ability of LNPs to be internalized by cardiomyocytes, three LNPs were chosen: the LNP that a) achieved the highest (LNP4), b) medium (LNP5), and c) lowest (LNP6) GFP expression in cardiomyocytes and formulated with fluorescent cholesterol. Then, the LNPs were incubated with cardiomyocytes at 0.2 μg of pDNA and fixed with 4% paraformaldehyde (PFA) at 2h, 4h, and 6h following transfection. Confocal images obtained at these time points confirmed the higher uptake of LNP4 over time by cardiomyocytes (Figure 4A).
To demonstrate the ionizable properties of LNPs, the pKa was checked using a TNS assay, as previously described.28 TNS assay revealed a pKa of 6.5 for LNP4, while LNP5 and LNP6 showed a pKa of 7.2 and 7.3, respectively (Figure 4B-D). To assess the effects of LNPs on cell viability, cardiomyocytes were then treated with LNPs 4–6, and after 48h, GFP fluorescence and cell viability were simultaneously measured. Data presented in Figure 4E, confirmed LNP4 as the top-performing LNP formulation from the mini library, displaying the highest GFP expression when compared to LNP5 and LNP6. Cell viability was higher than 90% for cardiomyocytes treated with LNP4 at doses of 0.4 and 0.8 μg of pDNA (Figure 4E). However, lower cell viability (75–85%) was observed for cardiomyocytes treated with LNP5 and LNP6 at doses of 0.4 and 0.8 μg of pDNA (Figure 4E). The stability of LNPs in serum conditions was assessed after incubation with BSA for 1, 3 and 12h at 37 °C. The size of LNPs was significantly increased after incubation with BSA compared to fresh LNPs over time (Figure S1A, Supplementary Material). However, it was observed only a slight increase (absolute value) in zeta potential of LNPs incubated with BSA compared to fresh LNPs (Figure S1B, Supplementary Material). The percentage of BSA adsorbed on LNP was approximately 10%, 18% and 31% after 1h, 3h and 12h of incubation, respectively (Figure S1C, Supplementary Material).
LNP4 Transfection Efficiency
To further determine the amount of pDNA that is needed to achieve the highest transfection efficiency, a dose–response curve was performed using the top-performing formulation LNP4 (0.00625–0.8 μg of pDNA). The maximum fluorescence was achieved using 0.2 μg of pDNA, with similar fluorescence intensity at higher doses of 0.4 and 0.8 μg of pDNA (Figure 5A). Next, to assess the fluorescence intensity over time, cardiomyocytes were transfected with LNP4 formulation (dose 0.2 μg of pDNA). The maximum fluorescence was achieved at day 4, which was kept over 8 days (Figure 5B and C). Moreover, the treatment with LNP4 (dose 0.2 μg of pDNA) achieved a transfection efficiency (GFP positive cells in the well plate) greater than 60% at day 2 and greater than 80% at day 4 (Figure 5D). Together, these results support LNP4 as the top formulation for pDNA delivery in cardiac cells and provide the optimized transfection methods for pDNA-LNPs in vitro.
In vivo Delivery of LNPs to Heart Tissue
Next, the in vivo delivery of the top-performing LNP4 was assessed, especially to the heart. LNP4 encapsulating pDNA were injected intravenously via tail vein injection into mice (Figure 6A). The heart was harvested and assessed by immunofluorescence 7 days post-injection. The LNP4-treated group was able to induce a twofold increase in GFP expression in the heart tissue compared to the control group (Figure 6B).
LNPs are Safe for pDNA Delivery to the Heart
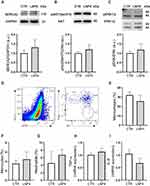
In vivo toxicity following LNP4 and PBS injection was assessed via analysis of myocytes morphology and contractility, cardiac function, inflammation induction, histopathological scoring, hematological parameters, and liver enzymes alanine aminotransferase (ALT) and aspartate aminotransferase (AST). Cardiomyocytes were isolated from previously treated mice to assess important parameters that reflect the cell function. For these experiments, ventricular myocytes were isolated and exposed acutely (5–10 min) to cell stress with isoproterenol (beta-adrenergic agonist) (Figure S2, Supplementary Material) and concomitantly increased the pacing rate (Figure S3A-C, Supplementary Material). Figure S2A-F and S4 (Supplementary Material) showed that LNP4 does not influence contractility function at the cellular level even with isoproterenol-induced stress or increased beating rate. Moreover, no alterations were observed in the cellular area, length, or width among the groups (Figure S2G-I, Supplementary Material), which together indicates that LNP4 treatment did not induce cellular remodeling. Cardiomyocyte function and survival were assessed via Western blotting. SERCA2, AKT, and ERK1/2 expressions were similar in mice treated with LNP4 and the control group (Figure 7A-C and Figure S4, Supplementary Material).
To investigate the potential of LNP4 to trigger inflammation in the heart, myeloid cell types were characterized via flow cytometry31 and inflammatory cytokines were assessed 7 days after treatment via qRT-PCR and compared to the control group. The myeloid cell population was characterized and subdivided into macrophages (from 32.08 ± 7.26 to 25.63 ± 13.93), monocytes (from 3.55 ± 1.40 to 4.47 ± 3.51), and neutrophils (from 4.54 ± 6.90 to 5.54 ± 0.94) (Figure 7D-G). No difference was found in the frequency of macrophages, monocytes, and neutrophils after treatment with LNP4. In addition, it was not found a difference in the expression of both inflammatory cytokines TNF-α, and IL-6 RNA (Figure 7H and I) indicating that the in vivo treatment with LNP4 was not able to trigger tissue inflammation.
Furthermore, inflammatory scoring of both heart and liver shows that LNP4 was well-tolerated at a dose of 10 µg, without causing any damage (Figure S5A-D, Supplementary Material). Negligible changes in counts of red blood cell (RBC), white blood cell (WBC), platelets, and lymphocytes levels were observed in mice treated with LNP4 and PBS (Table S1, Supplementary Material). In addition, there were no significant changes in ALT or AST levels in the serum of mice treated with LNP4 or PBS (Figure S5E, Supplementary Material), indicating that LNP4 enabled pDNA delivery to heart or liver without inducing enzyme release–associated toxicity at the time point evaluated here. Collectively, these results demonstrate that our LNPs do not induce heart damage, hematological disorder, or inflammatory response.
Discussion
Gene therapy is a promising approach to be applied in CVD, the leading cause of death worldwide.1,3 While significant progress continues to be made in gene therapy for the treatment of CVD, a key challenge remains in the method of delivery to the heart tissue.32 Viral vectors based on adenovirus and the adeno‐associated virus (AVV) have been extensively explored to deliver nucleic acid to the cardiovascular system. Significant progress using adenovirus has been made in pro-angiogenic gene therapy that targets vascular endothelial growth factor (VEGF) in the treatment of coronary heart disease,33–35 and peripheral vascular disease.36,37 In addition, gene therapy using viral vectors has also been applied to treat atherosclerosis, hyperlipidemias, peripheral ischemia, and arrhythmias.33,38–41 However, viral vectors stimulate inflammatory and immune responses, which raise safety concerns. Furthermore, the therapeutic efficacy of this approach remains a challenge in the heart tissue. Therefore, it is necessary to develop an efficient and safe gene delivery platform with the potential to be translated to the clinic.
In this study, non-viral vectors’ LNP formulations were developed and optimized, to deliver pDNA to cardiomyocytes, which are considered hard-to-transfect cells, and important cell types to study heart disorders.42 These LNPs are derivatives of prior materials with the ability to deliver mRNA and DNA, such as ionizable and other lipids.18 Ionizable lipid enables cellular uptake and escape of the endosomal compartment, cholesterol and DOPE provide stability to the lipid bilayer, and PEG-lipid conjugate prevents aggregation of vesicles and recognition by the mononuclear phagocytic system and also provides longer circulation.18 The literature has supported the potential of various ionizable lipids in mRNA delivery.17,18,31 Because of the structural differences between DNA and mRNA, improving DNA delivery requires further optimization in LNP formulation, which includes reformulating existing ionizable lipids. In this work, the novelty consists of developing pDNA-LNP formulations to enhance gene delivery to cardiomyocytes, by modulating nanoparticle lipid architecture, including the molar ratio of the lipid compositions, such as DOPE and cholesterol, and ionizable lipid:pDNA ratio. LNPs formulations with C12-200:pDNA ratio of 20:1 showed a slight increase in the encapsulation content as observed in previous studies as well as positive zeta potential values in contrast to LNPs with a ratio of 10:1.43 A rational explanation is the enhanced electrostatic complexation between the ionizable lipid C12-200 positively charged and negatively charged DNA molecule. As the DNA becomes more surrounded by the cationic lipid, there is an excess of positive charge, which reflects in the positive zeta potential values compared to the values of 10:1 formulations.44
By screening these unique LNPs formulations, a top-performing nanoparticle, termed LNP4, with enhanced transfection efficiency, and gene expression in cardiomyocytes was achieved. Taken that no significant increase in GFP expression was observed at 20:1 molar ratio of ionizable lipid:pDNA and to mitigate any concerns about possible lipid toxicity caused by increased lipid doses, LNP4 (10:1 C12-200:pDNA) was chosen as the top-performing formulation. In addition, a correlation between increased DOPE molar ratio with an enhanced GFP expression was observed. DOPE has been used in LNP formulations, as a helper lipid and associated with ionizable lipids, to improve transfection efficiency by destabilizing the endosomal membrane thereby facilitating the release of pDNA into the cytoplasm.43,45 In cationic liposomes, using 1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP), it has been reported that the in vitro transfection efficiency was mainly influenced by the DOTAP/DOPE ratio,46 which reinforces the important role of DOPE in the efficiency of LNPs. From these results, it was concluded that the composition of LNPs formulation is critical for pDNA delivery to cardiomyocytes.
As hypothesized, LNPs were able to deliver pDNA safely and effectively to cardiomyocytes. Together, our findings have broad implications for the advancement of basic research and cardiac gene therapies, as an efficient tool to understand the mechanisms of cardiac disorders, in terms of evaluating cardiac genes and recombinant protein expression.
Although the mechanism of uptake into cardiomyocytes remains to be determined, it was speculated that the enhanced GFP expression of LNP4 is related to improved uptake over time compared to other LNP formulations. Furthermore, it is well known that pKa of LNPs has a critical role in efficient nucleic acid delivery.47 As demonstrated by GFP expression, LNP4 formulation (pKa 6.5) has shown the highest transfection efficiency, which does not significantly increase at doses higher than 0.2 µg. Interestingly, the pKa of LNP4 is closer to endosomal pH compared to LNP5 and LNP6, which may have contributed to the increased uptake48 and consequently GFP expression. Thus, it was also speculated that the pKa of LNP4, closer to the endosomal pH, optimized the “proton-sponge” effect, endosomal disruption, and consequent release of pDNA to transcription and translation machinery. However, further studies are necessary to determine this mechanism and the traffic through the cytoplasm to the nucleus for gene expression.
The fact that GFP expression reached a plateau in doses higher than 0.2 µg could be related to the increased amount of PEG-lipid, which can reduce LNP interactions with target cells and limit transfection.49
For the experiment performed in vivo, a single intravenous injection was used to investigate the transfection efficiency and toxicity in vivo of the top-performing formulation. LNP4 was able to induce significant gene expression in the heart tissue. Further optimization starting from lead LNP4 formulation, including the use of barcoded DNA to screen LNPs in vivo and/or surface functionalization to enhance selectivity target to heart tissue will likely enhance the efficiency and avoid binding to plasma proteins as well as off-target effects. It is described that LNP administration directly in the lungs or via the intradermal route can trigger an inflammatory response increasing the frequency of neutrophils, which is considered a potent adjuvant.50 Recently, other studies have reported myocarditis in a small percentage of vaccinated people with mRNA vaccines, which uses LNPs as vectors.51 Therefore, the ability of our LNPs to trigger inflammation or other toxicity in the heart, such as cardiac function was also investigated. The levels of circulating myeloid cells and inflammatory cytokines were not significantly changed in the heart after LNP treatment. In addition, it was demonstrated that cell contractility and protein expression of SERCA2, AKT phosphorylation at Ser473, and pERK1/2, which reflect the cardiac cell function and survival, was not affected after LNP treatment. Collectively, our results have demonstrated that our LNPs work well in heart tissue and do not induce heart or liver damage, hematological disorder, or inflammatory response, which reproduces the safety of many ionizable LNPs that have been evaluated in animal models, and clinical trials in humans.52,53 Therefore, it is expected that future studies will yield high safety for other tissues, as well as the heart and liver.
Although more in vivo experiments are needed to assess biodistribution and to improve selectivity, we believe that the top-performing formulations described here may provide a basis for further optimization using other pDNAs as well as a platform for pDNA delivery for the treatment of CVD.
Conclusion
Here, it was presented evidence that LNP formulations were able to deliver pDNA to cardiomyocytes with enhanced gene expression. It was found that LNPs formulated with higher DOPE and lower cholesterol molar ratio exhibited enhanced delivery and GFP expression in cardiomyocytes. Our lead LNP was able to induce up to 80% of transfection efficiency in vitro and twofold higher fluorescence in vivo after 7 days of intravenous injection, with negligible toxicity. Further optimization, including functionalization of LNPs to enhance selectivity target to heart tissue, will likely enhance the efficiency and avoid off-target effects. Taken together, our results suggest the use of LNPs holds promise to improve pDNA delivery to cardiomyocytes.
Abbreviations
LNPs, lipid nanoparticles; CVD, cardiovascular diseases; NPs, nanoparticles; PEG, lipid-polyethylene glycol; GFP, Zsgreen a green fluorescent protein; DOPE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; ZP, zeta potential; HD, hydrodynamic diameter; PDI, polydispersity index; C14PEG, polyethylene glycol (PEG) 2000; EE, encapsulation efficiency; CTR, control; RBC, red blood cell; WBC, white blood cell; Cryo-TEM, cryogenic-transmission electron microscopy; NIH, National Institutes of Health; ICB-UFMG, Institute of Biological Sciences at Federal University of Minas Gerais; CAPI-UFMG, Center for Acquisition and Processing of Images; TNS, 6-(ptoluidinyl) naphthalene-2-sulfonic acid; ALT, alanine aminotransferase; AST, aspartate aminotransferase; DOTAP, 1,2-dioleoyl-3-trimethylammoniumpropane.
Ethics Approval and Informed Consent
Animal experimentation was carried out in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals, and approved by the Institutional Animal Care and Use Committee at UFMG (IACUC-UFMG 138/2018 and 177/2020).
Acknowledgments
This work was supported by CNPq (401390/2020-9; 442731/2020-5; Universal 2018), PRPq-UFMG, CAPES (88887.513270/2020-00), and FAPEMIG (RED-00282-16 Rede de Pesquisa e Inovação para Bioengenharia de Nanossistemas; Universal 2017, 2021 APQ-00826-21) for providing financial support for this investigation.
Disclosure
Dr Pedro Pires Goulart Guimaraes reports grants from the National Council for Scientific and Technological Development (CNPq), grants from CAPES, grants from FAPEMIG, during the conduct of the study. There are no other conflicts of interest to declare.
References
1. Virani SS, Alonso A, Aparicio HJ, et al. Heart disease and stroke statistics—2021 update. Circulation. 2021;143:8. doi:10.1161/CIR.0000000000000950
2. Yusuf S, Reddy S, Ôunpuu S, Anand S. Global burden of cardiovascular diseases. Circulation. 2001;104(22):2746–2753. doi:10.1161/hc4601.099487
3. World Health Organization. Cardiovascular diseases (CVDs); 2021. https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds).
4. Matkar PN, Leong-Poi H, Singh KK. Cardiac gene therapy: are we there yet? Gene Ther. 2016;23(8–9):635–648. doi:10.1038/gt.2016.43
5. Tarride JE, Lim M, DesMeules M, et al. A review of the cost of cardiovascular disease. Canadian J Cardiol. 2009;25(6):e195–e202. doi:10.1016/S0828-282X(09)70098-4
6. Tilemann L, Ishikawa K, Weber T, Hajjar RJ. Gene therapy for heart failure. Circ Res. 2012;110(5):777–793. doi:10.1161/CIRCRESAHA.111.252981
7. Boon RA, Dimmeler S. MicroRNAs in myocardial infarction. Nat Rev Cardiol. 2015;12(3):135–142. doi:10.1038/nrcardio.2014.207
8. Cannatà A, Ali H, Sinagra G, Giacca M. Gene therapy for the heart lessons learned and future perspectives. Circ Res. 2020;126(10):1394–1414. doi:10.1161/CIRCRESAHA.120.315855
9. Gabisonia K, Prosdocimo G, Aquaro GD, et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569(7756):418–422. doi:10.1038/s41586-019-1191-6
10. Lyon AR, Sato M, Hajjar RJ, Samulski RJ, Harding SE. Gene therapy: targeting the myocardium. Heart. 2008;94(1):89–99. doi:10.1136/hrt.2007.116483
11. Mason D, Chen YZ, Krishnan HV, Sant S. Cardiac gene therapy: recent advances and future directions. J Controlled Release. 2015;215:101–111. doi:10.1016/j.jconrel.2015.08.001
12. Sasano T, Kikuchi K, McDonald AD, Lai S, Donahue JK. Targeted high-efficiency, homogeneous myocardial gene transfer. J Mol Cell Cardiol. 2007;42(5):954–961. doi:10.1016/j.yjmcc.2007.02.004
13. Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. 2010;17(3):295–304. doi:10.1038/gt.2009.148
14. Ji J, Shang-Yi Y. Ultrasound-targeted transfection of tissue-type plasminogen activator gene carried by albumin nanoparticles to dog myocardium to prevent thrombosis after heart mechanical valve replacement. Int J Nanomedicine. 2012:2911. doi:10.2147/IJN.S32363
15. Sun L, Huang CW, Wu J, et al. The use of cationic microbubbles to improve ultrasound-targeted gene delivery to the ischemic myocardium. Biomaterials. 2013;34(8):2107–2116. doi:10.1016/j.biomaterials.2012.11.041
16. Suk JS, Xu Q, Kim N, Hanes J, Ensign LM. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv Drug Deliv Rev. 2016;99:28–51. doi:10.1016/j.addr.2015.09.012
17. Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–555. doi:10.1038/nrg3763
18. Guimaraes PPG, Zhang R, Spektor R, et al. Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J Controlled Release. 2019;316:404–417. doi:10.1016/j.jconrel.2019.10.028
19. Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Materials. 2021;6(12):1078–1094. doi:10.1038/s41578-021-00358-0
20. Bus T, Traeger A, Schubert US. The great escape: how cationic polyplexes overcome the endosomal barrier. J Materials Chem B. 2018;6(43):6904–6918. doi:10.1039/C8TB00967H
21. Yang S, May S. Release of cationic polymer-DNA complexes from the endosome: a theoretical investigation of the proton sponge hypothesis. J Chem Phys. 2008;129(18):185105. doi:10.1063/1.3009263
22. Cheng X, Lee RJ. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv Drug Deliv Rev. 2016;99:129–137. doi:10.1016/j.addr.2016.01.022
23. Granot Y, Peer D. Delivering the right message: challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics—An innate immune system standpoint. Semin Immunol. 2017;34:68–77. doi:10.1016/j.smim.2017.08.015
24. Mui BL, Tam YK, Jayaraman M, et al. Influence of Polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Therapy. 2013;2:e139. doi:10.1038/mtna.2013.66
25. Love KT, Mahon KP, Levins CG, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Nat Acad Sci. 2010;107(5):1864–1869. doi:10.1073/pnas.0910603106
26. Guatimosim S, Amaya MJ, Guerra MT, et al. Nuclear Ca2+ regulates cardiomyocyte function. Cell Calcium. 2008;44(2):230–242. doi:10.1016/j.ceca.2007.11.016
27. Otsu N, Threshold Selection A. Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern. 1979;9(1):62–66. doi:10.1109/TSMC.1979.4310076
28. Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Controlled Release. 2005;107(2):276–287. doi:10.1016/j.jconrel.2005.06.014
29. O’Brien J, Wilson I, Orton T, Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem. 2000;267(17):5421–5426. doi:10.1046/j.1432-1327.2000.01606.x
30. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
31. Krohn-Grimberghe M, Mitchell MJ, Schloss MJ, et al. Nanoparticle-encapsulated siRNAs for gene silencing in the haematopoietic stem-cell niche. Nat Biomed Eng. 2020;4(11):1076–1089. doi:10.1038/s41551-020-00623-7
32. Bass-Stringer S, Bernardo BC, May CN, Thomas CJ, Weeks KL, McMullen JR. Adeno-associated virus gene therapy: translational progress and future prospects in the treatment of heart failure. Heart Lung Circ. 2018;27(11):1285–1300. doi:10.1016/j.hlc.2018.03.005
33. Ylä-Herttuala S, Baker AH. Cardiovascular gene therapy: past, present, and future. Mol Therapy. 2017;25(5):1095–1106. doi:10.1016/j.ymthe.2017.03.027
34. Stewart DJ, Hilton JD, Arnold JMO, et al. Angiogenic gene therapy in patients with nonrevascularizable ischemic heart disease: a Phase 2 randomized, controlled trial of AdVEGF121 (AdVEGF121) versus maximum medical treatment. Gene Ther. 2006;13(21):1503–1511. doi:10.1038/sj.gt.3302802
35. Hedman M, Hartikainen J, Syvänne M, et al. Safety and feasibility of catheter-based local intracoronary vascular endothelial growth factor gene transfer in the prevention of postangioplasty and in-stent restenosis and in the treatment of chronic myocardial ischemia. Circulation. 2003;107(21):2677–2683. doi:10.1161/01.CIR.0000070540.80780.92
36. Flugelman MY, Halak M, Yoffe B, et al. Phase Ib safety, two-dose study of MultiGeneAngio in patients with chronic critical limb ischemia. Mol Therapy. 2017;25(3):816–825. doi:10.1016/j.ymthe.2016.12.019
37. Deev R. pCMV- vegf165 intramuscular gene transfer is an effective method of treatment for patients with chronic lower limb ischemia. J Cardiovasc Pharmacol Ther. 2015;20(5):473–482. doi:10.1177/1074248415574336
38. Kim Y, Zharkinbekov Z, Sarsenova M, Yeltay G, Saparov A. Recent advances in gene therapy for cardiac tissue regeneration. Int J Mol Sci. 2021;22(17):9206. doi:10.3390/ijms22179206
39. Ishikawa K, Weber T, Hajjar RJ. Human cardiac gene therapy. Circ Res. 2018;123(5):601–613. doi:10.1161/CIRCRESAHA.118.311587
40. Shimamura M, Nakagami H, Sanada F, Morishita R. Progress of gene therapy in cardiovascular disease. Hypertension. 2020;76(4):1038–1044. doi:10.1161/HYPERTENSIONAHA.120.14478
41. Bezzerides VJ, Prondzynski M, Carrier L, Pu WT. Gene therapy for inherited arrhythmias. Cardiovasc Res. 2020;116(9):1635–1650. doi:10.1093/cvr/cvaa107
42. Tonelli FMP, Lacerda SMSN, Paiva NCO, et al. Functionalized nanomaterials: are they effective to perform gene delivery to difficult-to-transfect cells with no cytotoxicity? Nanoscale. 2015;7(43):18036–18043. doi:10.1039/C5NR04173B
43. Kauffman KJ, Dorkin JR, Yang JH, et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15(11):7300–7306. doi:10.1021/acs.nanolett.5b02497
44. Ermilova I, Swenson J. DOPC versus DOPE as a helper lipid for gene-therapies: molecular dynamics simulations with DLin-MC3-DMA. Physical Chem. 2020;22(48):28256–28268. doi:10.1039/D0CP05111J
45. Du Z, Munye MM, Tagalakis AD, Manunta MDI, Hart SL. The role of the helper lipid on the DNA transfection efficiency of lipopolyplex formulations. Sci Rep. 2015;4(1):7107. doi:10.1038/srep07107
46. Kim BK, Hwang GB, Seu YB, Choi JS, Jin KS, Doh KO. DOTAP/DOPE ratio and cell type determine transfection efficiency with DOTAP-liposomes. Biochim Biophys Acta. 2015;1848(10):1996–2001. doi:10.1016/j.bbamem.2015.06.020
47. Jayaraman M, Ansell SM, Mui BL, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angewandte Chemie Int Edition. 2012;51(34):8529–8533. doi:10.1002/anie.201203263
48. Rennick JJ, Johnston APR, Parton RG. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat Nanotechnol. 2021;16(3):266–276. doi:10.1038/s41565-021-00858-8
49. Song LY, Ahkong QF, Rong Q, et al. Characterization of the inhibitory effect of PEG-lipid conjugates on the intracellular delivery of plasmid and antisense DNA mediated by cationic lipid liposomes. Biochim Biophys Acta. 2002;1558(1):1–13. doi:10.1016/S0005-2736(01)00399-6
50. Ndeupen S, Qin Z, Jacobsen S, Bouteau A, Estanbouli H, Igyártó BZ. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience. 2021;24(12):103479. doi:10.1016/j.isci.2021.103479
51. Verma AK, Lavine KJ, Lin CY. Myocarditis after Covid-19 mRNA vaccination. N Eng J Med. 2021;385(14):1332–1334. doi:10.1056/NEJMc2109975
52. Kulkarni JA, Myhre JL, Chen S, et al. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomedicine. 2017;13(4):1377–1387. doi:10.1016/j.nano.2016.12.014
53. Sabnis S, Kumarasinghe ES, Salerno T, et al. A Novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol Therapy. 2018;26(6):1509–1519. doi:10.1016/j.ymthe.2018.03.010
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.