Back to Journals » Infection and Drug Resistance » Volume 14
Phenotypic Detection of Carbapenem-Resistant Gram-Negative Bacilli from a Clinical Specimen in Sidama, Ethiopia: A Cross-Sectional Study
Authors Alemayehu T , Asnake S , Tadesse B, Azerefegn E, Mitiku E, Agegnehu A, Nigussie N, H/Mariam T, Desta M
Received 9 November 2020
Accepted for publication 13 January 2021
Published 2 February 2021 Volume 2021:14 Pages 369—380
DOI https://doi.org/10.2147/IDR.S289763
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Tsegaye Alemayehu,1 Solomon Asnake,1 Bereket Tadesse,2 Elshaday Azerefegn,2 Enkosilassie Mitiku,2 Asnakech Agegnehu,2 Netsanet Nigussie,2 Techilo H/Mariam,2 Moges Desta1
1Hawassa University College of Medicine and Health Science, School of Medical Laboratory Science, Hawassa, Ethiopia; 2Hawassa University Comprehensive Specialized Hospital Microbiology Laboratory, Hawassa, Ethiopia
Correspondence: Tsegaye Alemayehu
Hawassa University College of Medicine and Health Science, School of Medical Laboratory Science, P.O. Box 1560, Hawassa, Sidama, Ethiopia
Tel +251-91387241
Email [email protected]
Background: Carbapenem-resistant gram-negative bacteria are an emergent source of both community-acquired and healthcare-associated infection that poses a substantial hazard to public health. This study aimed to conclude the magnitude of carbapenem resistance gram-negative bacteria from a clinical specimen at Hawassa University Comprehensive Specialized Hospital.
Methods: A hospital-based cross-sectional study was accompanied from February 13 to June 7, 2020, in which consecutive patients with 103 gram-negative bacteria were encompassed. The isolates included were 54 urine, 17 blood, 17 pusses, 4 cerebrospinal fluid (CSF), 3 aspirates, 3 effusions, 2 stools, 2 ear discharges, and 1 nasal swab. A semi-structured questionnaire was used to gather socio-demographic data from the attendant and clinical data from the patient’s chart. Patients admitted in any wards and visited outpatients department were included for the study if gram-negative bacteria was identified for those who accepted the consent. A routine manual culture, Gram’s staining and biochemical tests used to identify the bacteria. Antibiotic susceptibility was determined for twelve antibiotics including cotrimoxazole, ceftazidime, meropenem, gentamycin, chloramphenicol, ampicillin, ciprofloxacin, cefotaxime, cefuroxime, nitrofurantoin, piperacillin-tazobactam, and amikacin using the Kirby-Bauer disc diffusion method. Modified carbapenem inactivation (mCIM) method was used to determine carbapenem resistance using meropenem disk as per the recommendation of Clinical and Laboratory Standards Institute guideline. Statistical package for social science software version 21 was used for data entry and analysis. The odds ratio at 95% confidence interval (CI) and p-value < 0.05 were taken as a statistically significant association.
Results: Generally, 111 gram-negative bacteria were identified from 103 patients. Of 111 isolates, thirteen isolates (nine resistance and four intermediates) were identified in disk diffusion testing for meropenem. Of this, 10 isolates were carbapenemases producer with the overall rates of 9% in the Modified carbapenem inactivation method (mCIM). Pseudomonas spp. 3 (30.0%), E. coli, K. pneumonia, Acinetobacter spp. each two (20.0%), and K. oxytoca 1 (10.0%) were identified as carbapenemases positive. The rates of the multidrug, extensive, pan drug were 86.5, 43.3, and 1.8, respectively. Ampicillin 94 (97.9%), followed by cefuroxime 52 (91.2%), cefotaxime 94 (88.7%), cotrimoxazole 58 (88.1%), ceftazidime 40 (83.3%), ciprofloxacin 47 (77.1%), nitrofurantoin 35 (70.0%), gentamycin 71 (65.7%), with high level of resistance. However, piperacillin-tazobactam 41 (48.8%), chloramphenicol 25 (47.2%), meropenem 13 (11.7%), and amikacin 9 (8.5%) were with low rates of resistance. In this study, there were no variables statically associated with carbapenem resistance that is p > 0.05.
Conclusion: Our study showed that carbapenem-resistant gram-negative bacilli are 9% in the study area. Our finding signposts that ampicillin, cefuroxime, cefotaxime, cotrimoxazole, ceftazidime, ciprofloxacin, nitrofurantoin, and gentamycin with a high rate of resistance > 50%. However, piperacillin-tazobactam, chloramphenicol, meropenem, and amikacin were at low rates of resistance. Therefore, a measure should be taken to contain carbapenem resistance gram-negative bacteria in the study area. Further, study with better method needs to be conducted to conclude the real scenario of carbapenem resistance.
Keywords: phenotypic, carbapenem resistance, gram-negative bacilli, clinical specimen, Hawassa, Ethiopia
Background
Carbapenem-resistant gram-negative bacteria (CR-GNB) are an evolving cause of both community-acquired and healthcare-associated infection (HCAI) that pose a significant threat to public health.1 Carbapenem is bactericidal β-lactam antimicrobials with demonstrated efficacy in severe infections caused by extended-spectrum β-lactamase (ESBL) producing bacteria.2 Drugs like meropenem, ertapenem, and imipenem are used to treat most of the infections affected by multidrug-resistant bacteria and can be castoff for the screening of carbapenem resistance in the bacteriology laboratory.3 Carbapenem resistance can be intrinsic or acquired by mutation or gene acquisition via horizontal gene transfer.4
Both the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) annually define the susceptibility breakpoints to commercially available carbapenem’s, including doripenem, ertapenem, imipenem, and meropenem for gram-negative species, although EUCAST no longer provides doripenem breakpoints.5,6 When a strain is found to be non-susceptible to carbapenem’s (ie, intermediate or resistant), the mechanism of resistance is still unknown.7–9 Thus, to confirm the production of carbapenemases and/or presence of other mechanisms, further biochemical assays and/or gene-based tests must be performed.6,7,10 The modified Carbapenem Inactivation Method (mCIM) is a simple phenotypic test that detects carbapenemases-producing gram-negative bacteria that have only been evaluated for use on bacterial colonies.11
Gram-negative bacteria, especially Enterobacteriaceae, are communal sources of both community-acquired and hospital-acquired infections, comprising urinary tract, bloodstream, and lower respiratory tract infections. These bacteria can gain genes encoding numerous antibiotic resistance mechanisms, containing ESBLs, AmpCs, and carbapenemases. β-Lactam drugs are regularly the key therapeutic choice for serious infections, and carbapenem, in particular, are frequently considered agents of last remedy.12 However, the rise of unusual b-lactamases with straight carbapenem-hydrolyzing action has contributed to an augmented prevalence of carbapenem-resistant Enterobacteriaceae (CRE). CRE is principally challenging assumed the occurrence with which Enterobacteriaceae causes infections.13
Klebsiella species and Escherichia coli (E. coli) are models of Enterobacteriaceae, a regular part of the human belly bacteria that can be converted carbapenem-resistant. Patients whose care involves strategies like ventilators (breathing machines), urinary (bladder) catheters, or intravenous (vein) catheters and patients who are taking extended courses of certain antibiotics are most at menace for CRE infections.14,15 Also, patients with comorbidities, ICU stay immunosuppression, the two extremities of age, and exposure to multiple antibiotics before the initial culture considered as risk factors for acquiring CR.16,17 Now a day, the medical effect of carbapenem resistance has become a public health problem around the world in terms of increased mortality, longer hospital stays, and costs of treatment.18
The frequency of CRE differs extensively between diverse class and different topographical areas. In the USA, carbapenem-resistance rates are revealed as 0.1% and 5.3% for E. coli and Klebsiella pneumoniae (K. pneumoniae), correspondingly, whereas in Europe, most nations report resistance rates below 1% for both pathogens.19 Even though studies from Africa stated a carbapenem resistance of (3.8–52.6%),20–23 still there are a few data on CR-GNB in Africa.21 While there are a few studies in Ethiopia conveyed on CR with a range of 2–27.1%,24–27 however, there is limited data in the study area. Therefore, this study intended to determine the magnitude of CR-GNB identified from clinical samples at Hawassa University Comprehensive Specialized Hospital (HUCSH).
Methods
Study Design, Area, and Period
A hospital-based cross-sectional study was accompanied at HUCSH since February 13 to June 7, 2020. The hospital is found in the capital city of Sidama at Hawassa, 275 kilometers (KMs) far from Addis Ababa, the capital city of Ethiopia. The altitude of the town is 1697m beyond sea level with a mean annual temperature and rainfall of 20.9 o C and 997.6 mL correspondingly. HUCSH was established in November 2005 and it serves about 12 million peoples. Patients looking for medical care obtain services at different outpatient and inpatient units (surgery, gynecology, and obstetrics, internal medicine, paediatrics, ophthalmology, psychiatry, radiology, pathology). The laboratory in the hospital analyzes arrays of tests, including parasitological, microbiological, immunological, hematological, and biochemical analysis. In the microbiology section, all aerobic culture and sensitivity testing done regularly from Monday to Sunday for 24 hours. The microbiology laboratory at HUCSH was established in 2010. It is the only laboratory in the region that participates for obtaining accreditation.
Population
All patients that visited the microbiology laboratory for routine culture and susceptibility testing through the study period were the source population. All patients confirmed with gram-negative isolates were the study population.
Eligibility
All patients that requested for culture and those agreed to join in the study included and patients refuse to participate in the study and patients with gram-positive isolates omitted from the study.
Variables of the Study
Carbapenem resistance is the dependent variable, whereas, age, sex, residence, wards, another hospital stays, length of hospital stay, sites of infection, an external device used, length of device indwelled, previous antibiotics usage, class of antibiotics, and underlined disease.
Sampling Technique
Convenient sampling technique employed to collect 103 consecutive patients with gram-negative isolates. The patient’s result and data were included if they are volunteering to contribute to the study.
Data Collection
Socio-Demographic and Clinical Data Collection
Trained data collectors used an organized questionnaire to gather socio-demographic as well as clinical data from patients and patient’s charts. If there is a positive growth for gram-negative bacteria in the laboratory the microbiologist inform the person in the reception and attach the questionnaires with laboratory report form. For those in cable patients and children, the attendant or caregivers were requested for participation, otherwise for the ambulatory patients directly interviewed for the socio-demographic data. The clinical data were collected from the patients’ chart and attending physician.
Sample Collection
All samples were collected based on the standard operating procedure (SOP) of HUCSH microbiology laboratory. Urine, puss, blood, ear discharge, eye swab, stool, cerebrospinal fluid (CSF), sputum and nasal swab sample suspected for any bacterial infection. The samples were sent from different wards. All samples were cultured on appropriate culture media. Samples collected for routine culture purposes were included if gram-negative bacteria were isolated.
Laboratory Diagnosis
Culturing
The samples were inoculated based on the essentiality of samples. Blood culture was collected with the sterile procedure and immediately inoculated to tryptone soy broth (TSB) at the site of collection. Then the bottle was transported for incubation in the microbiology laboratory. It is incubated at 37 C0 for five days. The bottle checked daily for the presence of growth indicators, ie, gas, pellicles, clot, and hemolysis. The sample subcultured blood agar plate (BAP), chocolate agar plate (CAP), and MacConkey agar (MAC) and gram stain were conducted at 24 hours incubation even if there were no growth indicators. If there were grown in solid media, identification performed based on their gram staining characteristics. Gram-negative organisms were included in this study. Finally, on day five, the bottle subcultured solid media if there were no growth reported as negative. Urine and puss were inoculated with BAP and MAC. Chocolate agar was included for ear discharge and nasal swab. Colony characteristics and gram staining are used as preliminary identification of isolates.
Biochemical Testing
Once the organism was identified as gram-negative in gram staining, serial biochemical testing that was prepared routinely performed to identify the isolates. Triple sugar iron agar, urea, citrate, mannitol fermentation, lysine iron agar, sulfur indole motility testing, and oxides used to distinguish the isolates to the species level.
Antibiotic’s Susceptibility Testing
The Kirby disk diffusion technique was used to perform the susceptibility testing on Muller Hinton agar (MHA).5 Twelve different antibiotics performed including cotrimoxazole - COT (1.25/23.75µg), ceftazidime - CAZ (30µg), meropenem - MER (10µg), gentamycin - GEN (10µg), chloramphenicol - CAF (30µg), ampicillin - AMP (10µg), ciprofloxacin - CIP (5µg), cefotaxime - CTX (30µg) cefuroxime - CRX (30µg), nitrofurantoin - NIT (5 µg), piperacillin-tazobactam - PIT (100/10 µg) and amikacin - AMK (30 µg). The recent clinical laboratory standard institute guideline as sensitive, intermediate, and resistant will interpret the result of measuring the zone of inhibition. Carbapenem resistance is determined by modifying the inactivation of carbapenem in which meropenem disk is used.
Modified Carbapenem Inactivation Method (mCIM)
Sterile inoculating loop, 1 μL for Enterobacteriaceae, and 10 μL for Pseudomonas and Acinetobacter spp. of test organisms were added into a tube having 2 mL of tryptic soy broth (TSB). The bacterial suspension was mixed for 10–15 seconds. Subsequently, a 10-μg MER disk was carefully added to the already prepared suspension. The suspension with disk was then incubated for 4 hours ± 15 minutes at 35°C ± 2°C in an ambient atmosphere. Just before the end of the 4-hour carbapenem inactivation step, a suspension of the mCIM quality control strain (E. coli ATCC 25,922) with turbidity equivalent to a 0.5 McFarland standard was arranged and the surface of an MHA dish inoculated using the technique for standard disk diffusion susceptibility testing. The meropenem disk then detached from the TSB suspension by a 10-μL inoculating loop disk employed onto the inoculated MHA plate, which then incubated in an upturned situation for 18–24 hours at 35°C ± 2°C in ambient air.5
mCIM Result Interpretation
The width of the zone of inhibition around each MEM disk was measured a zone diameter of 6–10 mm was taken to be a positive result (ie, carbapenemases production detected), 11–19 mm an indeterminate result and ≥20 mm a negative result (ie, No carbapenemases production detected). A thin ring of growth adjoining the MEM disk, representative of the leftover of the test organism from the TSB, was snubbed5 (Figure 1).
 |
Figure 1 Modified carbapenem inactivation method (mCIM) to determine carbapenem resistance of gram-negative bacteria from clinical specimens at tertiary care hospital, southern Ethiopia. |
Quality Control
Data quality warranted using standardized data gathering tools, pretesting of the questionnaires, appropriate training earlier the start of data assortment and rigorous supervision throughout data assemblage by the authors. For laboratory investigation, pre-analytical, analytical, and post-analytical phases of quality assurance amalgamated into Standard operating procedures (SOPs) of the microbiology laboratory stringently tracked. Besides, well-trained and skilled laboratory professionals participated in the laboratory investigation system. Media sterility checked after preparation by incubating for 24hrs. Quality control bacteria such as E. coli (ATCC-25,922), and P. aeruginosa (ATCC-27,853) found from the Ethiopian public health institute (EPHI) to check the characteristic of the colony while growing of respective media and biochemical tests.
Data Processing and Analysis
Data were managed using SPSS statistical software version 21 and presented in table and graph. The bivariate and multivariate logistic regression model used to check the real forecasters of a dependent variable. The odds ratio at 95% confidence level and p-value <0.05 was used for statistical significance.
Ethical Consideration
Hawassa University College of Medicine and Health Sciences institutional review board (IRB) has approved the proposal with a Ref. No: IRB/268/12. At that moment support letter was gained from the hospital management. Socio-demographic data and specimen were collected after written and/agreement gained from each child’s parents and patients. All information kept secret using codes and locking on the board. The result of the patient-reported to the clinician within three or four days and those who are culturally positive treated accordingly.
Operational Definitions
Multidrug-resistant bacteria (MDR): Bacteria that are resistant to at least one agent in three or more antibiotic categories.
Extensively drug-resistant (XDR) is non-susceptibility to at least one agent in all but two or fewer antibiotic categories (ie, bacterial isolates remain susceptible to only one or two categories).
Pan drug-resistant (PDR) is non-susceptibility to all agents in all antibiotic categories.28
Result
Socio-Demographic Characteristics
One-hundred three (103) patients with gram-negative isolates were involved in the study, of these 54 (52.4%) of them were male. Regarding age 62 (60.2%) was <5 years and the rest 40 (38.8%) was >6 years age group. The mean and standard deviation (SD) of the age for the study subject was 11.52 ± 16.2 years that range from 1 day to 70 years. Fifty-five (53.4%) of them were from urban and the rest 48 (46.6%) was a rural resident (Table 1).
Clinical Features and Associated Risk Factors
Patients enrolled in this study were from eight different wards that include paediatrics 38 (36.9%), surgical 16 (15.5%) neonatal intensive care units (NICU) 27 (26.2%), and others (medical, intensive care unit (ICU), ENT) 22 (21.4%). Thirty-two (31.1%) patients had another hospital stay and the rest 711 (68.9%) have no history of hospital stay. The length of hospital stay for <10 days was 64 (62.1%) and the rest 39 (37.9%) stay for >11 days that ranged (0–40 days). The mean and SD of hospital stay was 11.6 ± 9.12 days. The sites of infection identified from this study were 54 (52.4%) was a urinary tract infection and the rest 49 (47.6%) was from other sites of infection (bloodstream, surgical site, intestinal, respiratory, nervous system, soft tissue, otitis). Most of the study subjects 85 (82.5%) used one or more external devices during a hospital stay in which the mean and standard deviation of length of device usage was 14.7 ± 10.8 in the days that ranged from 1 to 60 days. Antibiotics were used by 92 (89.3%) of patients as treatment or prophylaxis before sample collection and the rest 11 (10.7%) of them were not taking on antibiotics. Of this was 70 (76.1%) used a single class of antibiotics and the rest 22 (23.9%) taken a combined class of antibiotics. Our study tried to perform a bivariate analysis for socio-demographic as well as for clinical data. There were no candidate variables for multivariable analysis in bivariate analysis p < 0.25. Therefore, there are no statically significant associated variables for carbapenem resistance p > 0.05 (Table 1).
Specimen Type
Of total 103 specimen, 54 (48.6%) was urine, 17 (15.3%) blood, 17 (15.3%) puss, 4 (3.6%) cerebrospinal fluid (CSF), 3 (2.7%) aspirates, 3 (2.7%) effusion, 2 (1.8%) stool, 2 (1.8%) ear discharge, and 1 (0.9%) nasal swab (Figure 2).
Frequency of Isolated Bacteria
Of 103 patients enrolled in the study, overall 111-gram negative bacteria were isolated. The utmost isolate was E. coli 34 (30.6%). Followed by K. pneumoniae 31 (27.9%), Acinetobacter spp. 11 (9.9%), Klebsiella oxytoca (K. oxytoca) 9(8.1%), P. aeruginosa 8 (7.2%), Klebsiella ozaenae (K. ozaenae) 6 (5.4%), Morganella morganii (M. morganii) 5 (4.5%), Klebsiella rhinoscolaris (K. rhinoscolaris), Citrobacter diversus (C. diversus) and Proteus mirabilis (P. mirabilis) 2 (1.8%) each, and Enterobacter agglomerus (E. agglomerus) 1 (0.9%) (Figure 3).
 |
Figure 3 Frequency of bacteria isolated for the study of carbapenem resistance of gram-negative bacteria from clinical specimens at tertiary care hospital, southern Ethiopia February 13–June 7, 2020. |
Antibiotic Resistance Patterns
The resistance patterns of antibiotics for each bacteria were determined accordingly, the most resisted antibiotic was ampicillin 94 (97.9%), followed by cefuroxime 52 (91.2%), cefotaxime 94 (88.7%), cotrimoxazole 58 (88.1%), ceftazidime 40 (83.3%), ciprofloxacin 47 (77.1%), nitrofurantoin 35 (70.0%), gentamycin 71 (65.7%), piperacillin-tazobactam 41 (48.8%), chloramphenicol 25 (47.2%), meropenem 13 (11.7%) and amikacin 9(8.5%) (Table 2).
The Magnitude of Multidrug Resistance
The level of multidrug resistance (MDR) checked for each isolate considering that bacteria that were resistant for three or more groups of antibiotics as MDR. Based on this definition, the level of MDR ranges from (0–100%). Pseudomonas spp., K. ozaenae, K. oxytoca was 100% MDR followed by K. pneumoniae 96.8%, E. coli 82.4%, Acinetobacter spp. 81.8%, M. morganii 80%, K. rhinoscolaris and C. diversus 50%, and the rest P. mirabilis and E. agglomerus with 0% (Table 3).
The overall MDR, XDR and PDR were 86.5%, 43.2%, 1.8%, respectively (Figure 4).
 |
Figure 4 Rates of MDR, XDR and PDR bacteria from clinical specimens at tertiary care hospital, southern Ethiopia from February 13 to June 7, 2020. |
The Magnitude of Carbapenem Resistance
In Kirby-disc diffusion techniques, 111 isolates were tested against meropenem disk, of these 13 isolates (nine bacteria were resistant and four isolates were intermediate). In modified carbapenem inactivation techniques 10 out of the 13 confirmed as carbapenem resistance with overall carbapenem resistance was 9.0% (3.6–14.6%) (Figure 5), of these isolates the predominant was Pseudomonas spp. 3(30.0%), followed by E. coli, K. pneumonia, Acinetobacter spp. each two (20.0%) and K. oxytoca 1(10.0%). All CR bacteria showed MDR, XDR, and one of the isolates was PDR.
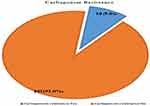 |
Figure 5 Magnitude of carbapenem resistance of gram-negative bacteria from clinical specimens at tertiary care hospital, southern Ethiopia from February 13 to June 7, 2020. |
Discussion
The arrival of resistance to carbapenem’s in gram-negative bacteria is a significant and increasing threat to public health in Africa.2 A load of infectious diseases is high in Africa,29 and gram-negative bacteria cause most of the common clinical infections such as UTI, septicemia, pneumonia, meningitis, and peritonitis. Years of deprived therapeutic antibiotic prescribing and lack of infection, prevention strategies in African hospitals have put patients in danger of acquiring these difficult to treat bacterial infections.30 The scourge of forged and substandard drug has only made the problem worse. Medical tourism has now resulted in patients acquiring these resistant bacteria in countries outside the continent and bringing them back to African hospitals.31 Our study conducted on all clinical specimen from different outpatient and inpatients sent for routine culture at HUCSH and only included for the study if gram negative bacteria isolated.
Our study shows that the overall CR-GNB was 9.0% (3.6% −14.6%). This is comparable to a study reported in India (14.6%).32 In contrast to our study, a higher rate of studies was reported from India (65%),33 (30%),34 Ethiopia (27.1%),35 and (25%),36 and lower prevalence studies reported from Ghana (2.9%),37 Addis Ababa, Ethiopia (2%),25 Germany (0.22%).38 This difference might be due to the study period, area, and laboratory method employed. Carbapenem is the last chance of treatment used in serious infections; however, due to the increment of carbapenem resistance bacteria, it is challenging for treating MDR, XDR, and PDR bacteria.25 In our study, all the isolates that are carbapenem resistant showed (100%) MDR, XDR, and (10%) PDR in which the other one isolate is indeterminate for carbapenemases.
The risk factors for the gaining of antibiotic-resistant bacterium demarcated as the antibiotic consumption and acquaintance, the use of the external device, healthcare exposure.39–41 Our study displays that CR-GNB is high among <5 years; however, it is not statistically significant (COR: 0.500, 95% CI: 0.096–2.609, p = 0.411). Similarly, a study from Nigeria reported as there is a difference in the proportion of age groups but not statistically associated.42 In study conducted in Northwest Ethiopia showed that being female (OR 4.46; P = 0.018), age (OR 1.08; P = 0.001), hospitalization (OR 5.23; P = 0.006), and prior antibiotic use (OR 3.98; P = 0.04) were associated risk factors for MDRE.42 Even though our study agrees as the rates of carbapenem resistance are high among age <5 years, with female sex and for those who had antibiotic usage before culture in general, but it is not statistically associated as p > 0.05. This difference might be explained with female patients are more susceptible for UTI as a natural phenomenon more than half of CR-GNB was isolated from UTI patients, ie, 5 out of 8 patients. The high rates of CR-GNB for those who had previous exposure of antibiotics may be explained that our hospital already started prescribing the carbapenem class of antibiotics and the bacteria may acquire these resistance genes. In studies elsewhere, patients with comorbidities, ICU stay, immunosuppression, the two extremities of age, and exposure to multiple antibiotics before the initial culture considered as risk factors for acquiring CR.17,18
In this finding, bacteria, which are CR, were Pseudomonas spp., Klebsiella spp., E. coli, and Acinetobacter spp. This is in agreement with studies testified in a systematic review from East Africa,43 India.34,44 This study identified as CR, three of four gram-negative organisms considered as ESKAPE, which are recognized as the most important emerging threats of this century that is Pseudomonas, Klebsiella, Acinetobacter, and Enterobacter.45,46
In this finding the overall MDR was 86.5% (79.1–91.0%), this value is comparable to a study reported from Ethiopia, in Bahir Dar (80.0%),47 Addis Ababa (81.5%),48 and Jimma (85%).49 In contrast to our study, a lower rate of MDR was reported from Debre Markos (72.2%),50 Bahir Dar (65.2%),51 Addis Ababa (68.3%),52 Northwest Ethiopia (54.3%),53 India (37.1%),54 (30%).34 However, a higher rate of MDR was reported from Gonder (92%),55 Addis Ababa (94.5%).56
Our study determined antibiotic susceptibility patterns of each isolate, based on this ampicillin are the most resisted antibiotics followed by cefuroxime, cefotaxime, cotrimoxazole, ceftazidime, ciprofloxacin, nitrofurantoin, and gentamycin, in descending order with greater than 50% of the resistance. In line with our study similarly a high resistance level of ampicillin was reported from Addis Ababa,35,56 Bahir Dar,53 Hawassa.57 In contrast to our study, a lower rate of resistance to ciprofloxacin and gentamycin was described,51,53,56 this designates that the resistance rates of commonly prescribed antibiotics getting a higher chance of resistance. In this study, lower rates of antibiotic resistance were observed for piperacillin-tazobactam, chloramphenicol, meropenem, and amikacin for all isolates in descending order, which is in line with reports from various parts of Ethiopia.53,58 This can be justified that the least prescribed antibiotics are still good but needs attention when introducing these drugs in the market as we are running out of effective antibiotics to cure the most vulnerable patients.
Conclusion
Our study showed that carbapenem-resistant gram-negative bacilli are 9% in the study area. The level of MDR is still alarmingly higher. Our finding signposts that ampicillin, cefuroxime, cefotaxime, cotrimoxazole, ceftazidime, ciprofloxacin, nitrofurantoin, and gentamycin with a high level of resistance >50%. However, piperacillin-tazobactam, chloramphenicol, meropenem, and amikacin were at low rates of resistance. In this study, there were no variables statically associated with carbapenem resistance. Therefore, a measure should be taken to decrease the increment of carbapenem resistance. The clinician should follow strictly the guideline for treating patients as we are running out of effective antibiotics. A study with a large sample of a gram-negative organism should be conducted for a better understanding of carbapenem resistance in the region. Finally, we recommend if empirical treatment is mandatory piperacillin-tazobactam, chloramphenicol, meropenem, and amikacin are more effective.
Limitation of the Study
- The drawback of this study is it only based on the phenotypic characteristic of carbapenem resistance that means not genotypic confirmed due to the budgetary issue and unavailability of setup.
- With this result, we could not generalize the carbapenem resistance in the region, as most of the patients come to this hospital after taking numerous treatments.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
Hawassa University College of Medicine and Health Sciences institutional review board (IRB) has approved the proposal (Ref. No. IRB/268/12). At that moment support letter was gained from the hospital management. Socio-demographic, clinical and laboratory data were collected after written informed consent and/agreement gained from each child’s parents and patients. All information kept secret using codes and locking on the board. The result of the patient-reported to the clinician within three or four days and those who are culturally positive treated accordingly. This study was conducted following the declaration of Helsinki.
Acknowledgments
First, we would like to thank Hawassa University for giving us this opportunity. Second, we would like to appreciate the staff of HUCSH Microbiology Laboratory for their support during analysis. Finally, we would give our heartfelt thanks to the study participants.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This study was funded by Hawassa University for data collection and laboratory analysis.
Disclosure
The authors report no conflicts of interest for this work.
References
1. WHO. Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities. 2017.
2. Codjoe FS, Eric S. Carbapenem resistance: a review. Med Sci. 2018;6(1):1. doi:10.3390/medsci6010001
3. Hu FCS, Xu X, Guo Y, Liu Y, Zhu D, Zhang Y. Emergence of carbapenem-resistant clinical Enterobacteriaceae isolates from a teaching hospital in Shanghai, China. J Med Microbiol. 2012;61(1):132–136. doi:10.1099/jmm.0.036483-0
4. Meletis G. Carbapenem resistance: an overview of the problem and future perspectives. Therap Adv Infect Dis. 2016;3(1):15–21. doi:10.1177/2049936115621709
5. CaLSI C. Performance Standards for Antimicrobial Susceptibility Testing. West Valley Road, Suite 2500, Wayne, Pennsylvania 19087 USA: Clinical and Laboratory Standards Institute; 2019:950.
6. Testing E Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0; 2019. Available from: http://wwweucastorg.
7. Bonomo RA, Burd EM, Conly J, et al. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis. 2018;66(8):1290–1297. doi:10.1093/cid/cix893
8. Gniadek TJ, Carroll KC, Simner PJ, Kraft CS. Carbapenem-resistant non-glucose-fermenting gram-negative bacilli: the missing piece to the puzzle. J Clin Microbiol. 2016;54(7):1700–1710. doi:10.1128/JCM.03264-15
9. Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18(9):1503–1507. doi:10.3201/eid1809.120355
10. Nordmann P, Poirel L. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2013;68(3):487–489. doi:10.1093/jac/dks426
11. Pierce VM, Simner PJ, Lonsway DR, et al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J Clin Microbiol. 2017;55(8):2321–2333. doi:10.1128/JCM.00193-17
12. LJaL B. The problem of carbapenemase-producing carbapenem-resistant Enterobacteriaceae detection. J Clin Microbiol. 2016;
13. Gupta NLB, Patel J, Kallen A, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53(1):60–67. doi:10.1093/cid/cir202
14. Ling MTY, Tan S, Amin I, How K, Tan K, Lee L. Risk factors for acquisition of carbapenem-resistant Enterobacteriaceae in an acute tertiary care hospital in Singapore. Antimicrob Resist Infect Control. 2015;4:26. doi:10.1186/s13756-015-0066-3
15. Kalam KQF, Baqi S, Baqi S, Baqi S, Baqi S. Risk factors for carbapenem-resistant bacteraemia and mortality due to gram-negative bacteraemia in a developing country. J Pak Med Assoc. 2014;64(5):530–536.
16. Meiling L, Wang J, Tan R, et al. Infection-prevention and control interventions to reduce colonisation and infection of intensive care unit-acquired carbapenem-resistant Klebsiella pneumoniae: a 4-year quasi-experimental before-and-after study. Antimicrob Resist Infect Control. 2019;8(8).
17. David Aguilera-Alonso LEG, Saavedra-Lozano J, Cercenado E, Fernando BA. Carbapenem-resistant gram-negative infections in children. Antimicrob Agents Chemother. 2019.
18. Johanna M, Vanegas OLP, Jiménez JN. Molecular epidemiology of carbapenem-resistant gram-negative bacilli from an infected pediatric population in tertiary-care hospitals in Medellín, Colombia: an increasing problem. BMC Infect Dis. 2016;16:463. doi:10.1186/s12879-016-1805-7
19. Nulens E, Gonzalo Bearman M, Fshea F. Guide to infection control in the hospital. Int Soc Infect Dis. 2018;1–16.
20. David O, Ogbolu LJVP, Mark AW. Opening Pandora’s box: high-level resistance to antibiotics of last resort in Gram 1 negative bacteria from Nigeria. bioRxiv. 2018. doi:10.1101/344093
21. Deogratius Okoche BBA, Katabazi FA, Kato L, Najjuka CF, Najjuka CF. Prevalence and characterization of carbapenem-resistant enterobacteriaceae isolated from Mulago National Referral Hospital, Uganda. PLoS One. 2015;10(8):e0135745. doi:10.1371/journal.pone.0135745
22. Igwe OA, Amauche Igwe H. Carbapenem-resistant Enterobacteriaceae (CRE) and gram-negative bacterial infections in south-west Nigeria: a retrospective epidemiological surveillance study</em>. AIMS Public Health. 2020;7(4):804–815.
23. Nada AB, Hisham AN, Kamal ME. Molecular characterization of carbapenem-resistant gram-negative bacteria in Khartoum (Sudan). Afr J Med Sci. 2020;5(2).
24. Setegn Eshetie CU, Gelaw A, Ayelign B, Endris M, Moges F. Multidrug-resistant and carbapenemase-producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrob Resist Infect Control. 2015;4(12).
25. Beyene DBA, Fantew S, Mihret A, Evans M, Evans M. Multidrug-resistant profile and prevalence of extended-spectrum β-lactamase and carbapenemase production in fermentative Gramnegative bacilli recovered from patients and specimens referred to National Reference Laboratory, Addis Ababa, Ethiopia. PLoS One. 2019;14(9):e0222911. doi:10.1371/journal.pone.0222911
26. Mitiku M, Ayenew Z, Desta K. Multi-drug resistant, extended spectrum beta-lactamase and carbapenemase producing bacterial isolate among septicemia suspected under five children in Tikur Anbesa Specialized Hospital, Addis Ababa Ethiopia: cross-sectional study. 2019.
27. Gashaw M, Berhane M, Bekele S, et al. Emergence of high drug-resistant bacterial isolates from patients with healthcare-associated infections at Jimma University medical centre: a cross-sectional study. Antimicrob Resist Infect Control. 2018;7(1):138.
28. Magiorakos APSA, Carey RB, Carmeli Y, Falagas ME, Giske CG. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
29. Fenollar F, Mediannikov O. Emerging infectious diseases in Africa in the 21st century. N Microbes N Infect. 2018;26:S10–S18. doi:10.1016/j.nmni.2018.09.004
30. Llor C, Bjerrum L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf. 2014;5(6):229–241. doi:10.1177/2042098614554919
31. Aliyu S. Carbapenemase-producing Gram-negative bacteria: an emerging threat to health care in Africa. Ann Niger Med. 2013;7(1):1. doi:10.4103/0331-3131.119978
32. Kaur MGS, Kaur T. Detection of carbapenem-resistant gram-negative bacteria in clinical isolates from a tertiary care hospital. J Bacteriol Mycol Open Access. 2016;2(1):00011.
33. Devi P. Incidence of carbapenem-resistant nonfermenting gram-negative bacilli from patients with respiratory tract infections among intensive care units. Int J Res Med Sci. 2017;3(6):4.
34. Mate PDK, Devi K, Damrolien S, Devi P. Prevalence of carbapenem resistance among Gram-negative bacteria in a tertiary care hospital in north-east India. IOSR J Dent Med Sci. 2014;13(12):56–60. doi:10.9790/0853-131235660
35. Tadesse MZ, Kassu D. Multi-drug resistant, extended-spectrum beta-lactamase and carbapenemase-producing bacterial isolates among children under five years old suspected bloodstream infection in a specialized Hospital in Ethiopia: cross-sectional study. 2019.
36. Gashaw MBM, Bekele S, Kibru G, et al. Emergence of high drug-resistant bacterial isolates from patients with healthcare-associated infections at Jimma University medical centre: a cross-sectional study. Antimicrob Resist Infect Control. 2018;7:138.
37. Codjoe FDE, Smith T, Miller K, Miller K. Phenotypic and genotypic characterization of carbapenem-resistant gram-negative bacilli pathogens from hospitals in Ghana. Microbial Drug Resist. 2019;25(10):1449–1457. doi:10.1089/mdr.2018.0278
38. Katchanov JAL, Klupp E, Both A, et al. Carbapenem-resistant Gram-negative pathogens in a German university medical centre: prevalence, clinical implications and the role of novel β-lactam/β-lactamase inhibitor combinations. PLoS One. 2018;13(4):e0195757. doi:10.1371/journal.pone.0195757
39. Souli M, Galani I, Giamarellou H. Emergence of extensively drug-resistant and pan drug-resistant Gram-negative bacilli in Europe. Eurosurveillance. 2008;13(47):19045.
40. Brink A, Coetzee J, Clay C, et al. The spread of carbapenem-resistant Enterobacteriaceae in South Africa: risk factors for acquisition and prevention. SAMJ. 2012;102(7):599–601. doi:10.7196/SAMJ.5789
41. Codjoe FS. Detection and Characterisation of Carbapenem-Resistant Gram-Negative Bacilli Infections in Ghana. Sheffield Hallam University; 2016.
42. Adesanya OA, Igwe HA. Carbapenem-resistant Enterobacteriaceae (CRE) and gram-negative bacterial infections in south-west Nigeria: a retrospective epidemiological surveillance study. AIMS Public Health. 2020;7(4):804.
43. Kenneth SDK, Edward W, Francis E, Ejobi F. A systematic review: the current status of carbapenem resistance in East Africa. BMC Res Notes. 2018;11:629. doi:10.1186/s13104-018-3738-2
44. ASaN K Prevalence and susceptibility analysis of carbapenem-resistant gram-negative pathogens in Tertiary Care Hospital, Mumbai; 2020. Available from: https://wwwjbclinpharmorg.
45. Peterson L. Bad bugs, no drugs: no ESCAPE revisited. Clin Infect Dis. 2009;49:992–993. doi:10.1086/605539
46. Souha SZA, Kanafani ZA. Current concepts in antimicrobial therapy against resistant gram-negative organisms: extended-spectrum β-lactamase–producing enterobacteriaceae, carbapenem-resistant enterobacteriaceae, and multidrug-resistant Pseudomonas aeruginosa. Mayo Clin Proc. 2011;86(3):250–259. doi:10.4065/mcp.2010.0674
47. Feleke MSE, Wondwossen A, Feleke M, et al. High prevalence of extended-spectrum beta-lactamase-producing Gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS One. 2019;14(4):1–13.
48. Bitew A. High prevalence of multi-drug resistance and extended spectrum beta-lactamase production in non-fermenting gram-negative bacilli in Ethiopia. Infect Dis. 2019;12:1–7.
49. Mohammedaman MAA, Tsegaye S, Kim H. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13(14):3–10. doi:10.1186/1476-0711-13-3
50. Wondemagegn MBA, Mulat Y, Tadesse H, Haimanot A, Dereje A, Abate D. Bacterial agents and antibiotic resistance profiles of infections from different sites that occurred among patients at Debre Markos Referral Hospital, Ethiopia: a cross‑sectional study. BMC Res Notes. 2017;10:254. doi:10.1186/s13104-017-2584-y
51. Mulugeta KBA. Bacteriology and antibiogram of pathogens from wound infections at Dessie Laboratory, North-East Ethiopia. Tanzan J Health Res. 2011;13(4).
52. Dejenie SAA, Melese H, Tesfaye L, Hiwot K, Kassu D. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control. 2019;8(39):3–12. doi:10.1186/s13756-018-0456-4
53. Derese HAD, Daniel M, Yohannes Z, Yesuf A, Seble W, Fantahun B. Drug resistance patterns of bacterial isolates from infected wounds at Bahir Dar Regional Health Research Laboratory Center, Northwest Ethiopia. Ethiop J Health Dev. 2016;30(3):113–117.
54. Silpi BPS, Monali R. Multidrug-resistant and extensively drug-resistant bacteria: a study. J Pathog. 2015;2016:5.
55. Abtie HT, Teshome B, Gebreselassie D, Mihiretie GD. The bacterial profile and antibiotic susceptibility pattern among patients with suspected bloodstream infections, Gondar, north-west Ethiopia. Pathol Labor Med Int. 2018;10:1–7. doi:10.2147/PLMI.S153444
56. Degefu AB, Surafel F, Amete M, Martin E. Multidrug-resistant profile and prevalence of extended-spectrum β-lactamase and carbapenemase production in fermentative Gram-negative bacilli recovered from patients and specimens referred to National Reference Laboratory, Addis Ababa, Ethiopia. PLoS One. 2019;14(9):1–13.
57. Tsegaye AMA, Enkosilassie M, Mengistu H. The burden of antimicrobial resistance at tertiary care hospital, southern Ethiopia: a three years’ retrospective study. BMC Infect Dis. 2019;20(585):1–8.
58. Aynalem ME, Teklay G, Moges T, Feleke M. Bacterial isolates and their antimicrobial susceptibility patterns of wound infections among inpatients and outpatients attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int J Microbiol. 2017;2017:10.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




