Back to Journals » Infection and Drug Resistance » Volume 13
Elevated Level of Imipenem-Resistant Gram-Negative Bacteria Isolated from Patients Attending Health Centers in North Gondar, Ethiopia
Authors Abda EM , Adugna Z, Assefa A
Received 22 October 2020
Accepted for publication 3 December 2020
Published 17 December 2020 Volume 2020:13 Pages 4509—4517
DOI https://doi.org/10.2147/IDR.S287700
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Ebrahim M Abda,1 Zenebe Adugna,2 Adugna Assefa2
1Department of Biotechnology, College of Biological and Chemical Engineering, Addis Ababa Science and Technology University, Addis Ababa, Ethiopia; 2Department of Biology, College of Natural and Computational Sciences, University of Gondar, Gondar, Ethiopia
Correspondence:Ebrahim M Abda
Department of Biotechnology, College of Biological and Chemical Engineering, Addis Ababa Science and Technology University, P.O. Box.16417, Addis Ababa, Ethiopia
Tel +251966917654
Email [email protected]
Background: The frequent identification of resistant bacteria in hospitals constantly presents antimicrobial therapy with a challenge. Imipenem, once considered an extremely powerful antibiotic against multidrug-resistant bacterial infections, is losing its effectiveness. Its use in empirical therapy with inadequate or nonexistent antimicrobial stewardship programs has further triggered bacterial resistance in low-income countries. Therefore, this study aimed at identifying imipenem-resistant Gram-negative bacteria from patients who were referred to health centers in North Gondar, Ethiopia.
Methods: A total of 153 sputum samples were used to isolate Gram-negative bacteria. The isolates, which were resistant to imipenem, were identified by standard biochemical tests and 16S rRNA sequencing. The Kirby-Bauer disk diffusion method was used to determine the sensitivity or resistance of the isolate to diverse antimicrobial agents.
Results: The study identified 79 imipenem-resistant bacterial isolates from eight genera with clinically relevant microorganisms, including Acinetobacter baumannii (20.77%), Klebsiella pneumoniae (19.48%), Pseudomonas aeruginosa (16.88%), and Serratia marcescens (14.28%). Overall, imipenem-resistant bacterial isolates were detected in 31 samples (20.26%). Additionally, a remarkably high level of resistance to most antibiotics was observed among isolates of Klebsiella pneumoniae and Acinetobacter baumannii. Gentamycin is the most active antibiotic against many of the isolates, while β-lactams appear to be less effective.
Conclusion: The study indicated that many Gram-negative bacteria were resistant to imipenem with parallel resistances to other antimicrobials. Hence, the prescription of imipenem within the region should be according to the antibiotic resistance profiles of the multi-drug resistant bacteria.
Keywords: Gram-negative bacteria, imipenem, North Gondar, 16S rRNA sequencing
Background
Resistance to antimicrobial agents is one of the main problems in modern medicine. Annually, 33,000 people in the EU and 35,000 people in the US die from antibiotic-resistant infections.1,2 Indeed, the burden of antimicrobial resistance (AMR) on economic growth is much greater in low-income countries, such as Ethiopia, than wealthy countries.3,4
In low-income countries, AMR often occurs with antimicrobial agents prescribed to treat respiratory infections, diarrhea, sexually transmitted diseases, and malaria.5,6 Among the well-known leading causes of morbidity and mortality in Ethiopia include respiratory infections caused by diverse etiological agents, notably Streptococcus pneumonia and Staphylococcus aureus, as well as many Gram-negative bacteria.7,8 These etiological agents are becoming increasingly resistant to a number of antimicrobial agents, making the treatment of respiratory infections difficult and expensive.
Carbapenems are considered the last line of defense against bacterial multidrug-resistant (MDR) infections and have lately been introduced to antimicrobial therapy, particularly in low-income countries due to their higher prices. Imipenem is one of the carbapenem that is used to treat respiratory infections.9,10 Nevertheless, increased resistance to imipenem has been documented worldwide in parallel with its application. Additionally, Gram-negative bacteria impose more risk with respect to AMR owing to the presence of MDR pumps, plasmids harboring antibiotic resistance genes, and various gene transfer mechanisms that play a role in the acquisition of resistance genes. With regard to Ethiopia, factors such as insufficient surveillance, limited diagnostic laboratories, and resource constraints have limited resistance profiling of the etiological agents responsible for leading cause of hospitalization.11,12
Therefore, this study aimed to identify imipenem-resistant Gram-negative bacteria from patients who were referred to the Gondar University referral hospital and the Maksegnit health center. Additionally, drug resistance profiling of these bacteria was examined, which could be beneficial for the selection of antimicrobial agents for local empirical therapy.
Materials and Methods
Sample Collection, Handling, and Processing
Sputum samples were collected from patients who visited the Gondar University referral hospital and the Maksegnit health center from March 2019 to June 2019. A total of 153 sputum samples were collected from patients with productive cough and dyspnea. The standard procedures were generally applied for collection and analysis. Briefly, patients were instructed to rinse their mouths with water, and then a sterile wide-mouth container was used to collect sputum. Sputum specimens with much watery saliva were excluded from the analysis. Thereby, only purulent sputum specimens were transported to the microbiology laboratory with cold transportation equipment. Additionally, all analyzed samples contained at least 25 polymorph nuclear leukocytes and 10 epithelial cells.
Culturing, Bacterial Isolation, and Biochemical Test
Initially, all culturable Gram-negative bacterial species were cultured by plating aliquots of the sputum specimens on MacConkey agar. The plates were incubated at 37°C for 24 h, and if no visible colonies were observed, the incubation period was extended to 48 h. All samples with positive cultures were transferred to Muller-Hinton agar plates and then impregnated with imipenem disc (10 µg) following the standard antibiotic susceptibility testing procedures. To identify the imipenem-resistant bacteria, the measured zone of inhibition was interpreted according to the standard and the interpretive criteria described in the Clinical Laboratory Standards Institute (CLSI) at the time of the tests.13 All imipenem-resistant bacteria were further characterized biochemically, including oxidase, catalase, indole, hydrogen sulfide, methyl red, Voges-Proskauer, lysine decarboxylase, glucose acidification, gelatin hydrolysis, motility, urea, citrate, growth at 42°C and fermentation of sugars in Triple sugar iron agar (TSI).14 Final confirmation of bacterial isolates was done by a molecular approach using the 16S rRNA gene sequencing method. Finally, lipase and protease production, and motility patterns were assayed as described in.15,16
16S rRNA Sequencing
The DNA was extracted by a boiling method.17 Briefly, colonies grown on LB agar plates were diluted in 40 μL Tris EDTA (TE) buffer and heated for 10 min at 55°C followed by 10 min heating at 80°C. About 80 μL TE was added to the boiled suspension before centrifugation at 6000rpm for 5 min. The supernatant was then transferred to clean Eppendorf cups and stored at −20°C or used immediately. For routine polymerase chain reaction (PCR), about 2–5 μL of the suspension was used.
PCR amplifications were performed using a thermal cycler (Bio-Rad Laboratories) by employing two sets of previously published primers (synthesized by Eurofins MWG GmbH, Ebensburg, Germany): 616V 5′-AGAGTTTGAT(CT)(AC)TGGCTCAG-3′ and 1492R 5′-CGG(CT)TACCT TTTACGAG-3′.18,19 An alternative forward primer GM1F 5′-CCAGCGGCCGCGGTGGTAAT-3′20 was used with 1492R reverse primer on the condition that the other primer set was failed to generate PCR products. The standard PCR reaction conditions were initial denaturation at 95°C for 7 min, followed by 35 cycles of 30 s denaturation at 95°C, annealing at 50°C for 30 s, extension at 72°C for 90 s, and a final extension at 72°C for 5 min. The PCR products were evaluated by DNA electrophoresis and visualized and documented using a UV light gel documentation device (Bio-Rad Laboratories, Munich, Germany). PCR primers were used to sequence the purified PCR fragments under conditions recommended by the manufacturer (Eurofins MWG GmbH, Ebensburg, Germany).
Antibiograms
Antimicrobial sensitivity tests were performed using the Kirby-Bauer disc diffusion method on Muller-Hinton agar by following the CLIS.13 Isolated Gram-negative bacteria were tested against imipenem (10 µg), gentamycin (10 μg), trimethoprim/sulfamethoxazole (25 μg), ampicillin (25 μg), chloramphenicol (30 μg), amoxycillin/clavulanic acid (30 μg), norfloxacin (10 μg) and tetracycline (30 μg) and all were Oxoid (Basingstoke, Hampshire, England).
Quality Control
All culture media were prepared according to the manufacturer’s instructions. The batch of the prepared media was checked for sterility by incubating samples of the plate at 37°C for 24 h. Moreover, E. coli ATCC (25922) and Pseudomonas aeruginosa ATCC (27853) were used as a standard strains for quality control.
Ethical Considerations
Protocol for patient participation in the study followed the principles of the Declaration of Helsinki and was approved by the Ethical Committee of the University of Gondar, College of Natural and Computational Sciences. Official permission from concerned offices and study participants were also obtained. Data were collected after obtaining informed consent from study participants. All the information obtained from study participants were kept confidential.
Results
Isolation of Gram-Negative Bacteria Resisting Higher Generation of β-lactam
In this study, a total of 153 sputum samples were examined for culturable Gram-negative bacteria. Most (74%) of the samples were collected from the University of Gondar referral hospital and the rest from the Maksegnit health center. Out of 153 samples, 110 (72%) had Gram-negative bacteria, while 43 (28%) had undetectable growth (Figure 1). On the basis of reduced susceptibility to imipenem disc (10 µg), imipenem-resistant bacterial isolates were detected in 31 samples (20.26%). Of these, 22.5% of the samples from the Maksegnit health center showed resistance to imipenem, while only 19.46% for the University hospital (Table 1). Overall, 79 imipenem-resistant bacterial isolates were obtained from 153 patients.
 |
Table 1 Gram-Negative Bacteria Demonstrating Imipenem Resistance Isolated from the Two Health Centers in North Gondar, Ethiopia |
 |
Figure 1 Percentage of culture positive and negative samples on MacConkey agar plates after 24 hours of growth. |
Diversity of Gram-Negative Bacteria in Sputum Specimens
To identify the diversity of bacteria, conventional biochemical tests with 16S rRNA gene sequencing was carried out for imipenem-resistant isolates. Sequence analysis of the PCR-amplified 16S rRNA gene indicated that many Gram-negative bacteria of medical importance were recovered from the clinical samples (Figure 2). Eight genera from both health centers were identified, including Acinetobacter, Serratia, Klebsiella, Pseudomonas, Stenotrophomonas, Enterobacter, Escherichia, and Kluyvera. More importantly, Acinetobacter baumannii (20.77%), Klebsiella pneumoniae (19.48%), Pseudomonas aeruginosa (16.88%), and Serratia marcescens (14.28%) were isolated more frequently in samples that were positive for imipenem resistance. It is noteworthy that most of the clinically relevant Acinetobacter species, including A. baumannii, Acinetobacter pittii, Acinetobacter junii, and Acinetobacter calcoaceticus, were identified in sputum samples using a number of distinctive biochemical tests, including glucose acidification, growth at 42°C and gelatin hydrolysis. Additionally, imipenem-resistant Enterobacter hormaechei (3.89%), Stenotrophomonas maltophilia (3.89%), and Kluyvera cryocrescens (2.59%) were present in fewer samples (Figure 2).
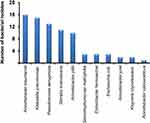 |
Figure 2 Diversity of Gram-negative imipenem-resistant bacteria identified by biochemical studies and 16S rRNA sequencing. |
Extracellular Enzyme Production and Motility Patterns of Emerging Nosocomial Pathogen Stenotrophomonas maltophilia
From our isolates, it was reported that P. aeruginosa has a cooperative pathogenicity with S. maltophilia.21 This affects the progression of the disease and also the choice of treatment, since one strain of these bacteria offers the other strain protection against some antimicrobial agents. Thereby, the identification of S. maltophilia from sputum samples has further motivated us to investigate the virulence properties of these isolates by analyzing motility and production of extracellular enzymes. Lipase and protease were secreted from all isolates, while the motility patterns were different. S. maltophilia undergoes three types of motility: twitching, swarming, and swimming. Motility patterns were assayed by growing the isolates overnight and recording their ability to diffuse in soft agar plates. Therefore, two of our clinical isolates showed twitching motility weakly, while no isolate had strong and moderate twitching motility (Figure 3). When isolates were investigated for swarming motility, the result showed that one isolate was strong, the other moderate, and the third weak. Finally, two of the S. maltophilia isolates demonstrated strong swimming motility patterns (Figure 3).
Antimicrobial Susceptibility Profiles of the Isolated Gram-Negative Bacteria
Antimicrobial susceptibility profiles revealed that our clinical isolates were most resistant to β-lactams, but most susceptible to aminoglycoside antibiotic-gentamycin (Table 2). In particular, isolates of A. baumannii showed complete resistance to ampicillin, amoxycillin/clavulanic acid, and trimethoprim/sulfamethoxazole, while the resistance to gentamycin was noticeably observed in 25% of the isolates. Moreover, 30% of the tested A. pittii isolates demonstrated resistance to chloramphenicol (Table 2).
 |
Table 2 Antimicrobial Resistance Pattern of Imipenem-Resistant Gram-Negative Bacteria Recovered from Sputum Samples |
The results of antimicrobial susceptibility testing also showed that 86.67%, 93.33%, and 100% of the tested K. pneumoniae isolates were resistant to amoxycillin/clavulanic acid, trimethoprim/sulfamethoxazole, and ampicillin, respectively (Table 2). Uncommon pathogens, including A. junii, K. cryocrescens, and A. calcoaceticus were mainly resistant to ampicillin and amoxycillin/clavulanic acid.
Moreover, many of the imipenem-resistant isolates also showed resistance to three and more classes of antimicrobial agents (multi-drug resistant). Specifically, 16 (100%) of A. baumannii, 15 (100%) of K. pneumoniae, 12 (92.30%) of P. aeruginosa, 11 (100%) of S. marcescens, 3 (100%) of E. coli and 2 (100%) of K. cryocrescens isolates were found MDR (Table 3). Clinical isolates of E. hormaechei, on the other hand, were broadly susceptible to wide-range of antibiotics (Table 3).
 |
Table 3 Multidrug-Resistance Profiles of Imipenem-Resistant Gram-Negative Bacteria Recovered from Sputum Samples |
Discussion
The healthcare system in low-income countries faces several problems. Resistance to antimicrobial agents further worsens the condition, affecting the limited resources and infrastructure of many hospitals. In the current study, 72% of sputum samples from patients who visited two health centers were positive for culturable Gram-negative bacteria. Isolation frequency between 42 and 83% has been documented in numerous health centers in Ethiopia and elsewhere in low-income countries.22–24 Also, a substantial difference in the isolation rate exists between high- and low-income countries, and higher rates from numerous clinical samples were common in countries of the latter category,25 which is consistent with the data presented in this study.
In our study, imipenem-resistant bacterial isolates were detected in 20.26% of the samples. Moreover, imipenem-resistant bacteria were identified at a slightly higher rate at the Maksegnit health center, indicating a rapid evolution of AMR in rural health clinics in addition to hospitals. Given the recent introduction of imipenem to the country’s treatment strategy,26 this implied increased detection of imipenem-resistant bacteria, which could be linked to many factors.27,28 In fact, the foremost important finding of this study could partly be attributed to the increased administration of imipenem for both documented infections and empirical therapy. In similar studies conducted previously, carbapenem-resistant pathogens occurred with a doubling of the carbapenem prescription in Germany.28 Another study conducted in France described that exposure to imipenem was primarily responsible for the development of several imipenem-resistant Gram-negative bacilli.29 In Africa, the level of resistance varied considerably between countries, with Tanzania having the highest carbapenem resistance at 35%, while the Democratic Republic of the Congo had the lowest level at 0.96%.30 Among the carbapenem antibiotics, resistance to imipenem was reported in 4% of the isolate from various clinical samples in Ghana.31 Moreover, an earlier study reported a 12.2% prevalence of carbapenemase-producing Enterobacteriaceae among Ethiopian children.32
The predominant imipenem-resistant bacteria identified in our study included A. baumannii, S. marcescens, K. pneumoniae, and P. aeruginosa. A similar group of bacteria was reported by Cadjoe and Dankor in their study, according to which K. pneumoniae, P. aeruginosa, and A. baumannii were described as the most important carbapenem-resistant bacterial pathogens.33 Additional studies also reported increased isolation of these bacteria in wounds,34 urine,35 and various clinical specimens.36
Many of these pathogens have been already implicated in nosocomial and community-acquired bacterial infection. K. pneumoniae is the principal cause of respiratory infections in Ethiopia and is also constantly identified from sputum samples.36 Although respiratory infections by the A. baumannii and S. marcescens are less frequent, their strong isolation might be linked to the event of resistance to imipenem. Additionally, S. maltophilia, a pathogen mainly associated with infection of immunocompromised persons, was recovered from the sputum sample with less frequency (2%). In fact, earlier studies reported different isolation percentage, which was 0.13% (blood), 0.03% (urine), and 4% (sputum).15 Although the microbe is primarily responsible for the infection of patients with cystic fibrosis (CF), the examined health centers still lack facilities for treating CF patients. However, due to its virulence potential (protease and lipase, Figure 3 and16) and the previously reported cooperative pathogenicity21 with the predominantly isolated bacteria (P. aeruginosa) in the study area, it could affect the progression of the disease in patients and thereby influence therapeutic choices.
Many of the Gram-negative bacteria implicated within the current study demonstrated multiple drug resistance, especially high resistance to ampicillin, amoxycillin/clavulanic acid and trimethoprim/sulfamethoxazole. The high-level of resistance to β-lactams might be attributed to an extended-spectrum β-lactamases (ESBL), metalloenzyme β-lactamases, which are mediated by other intrinsic or by transferable carbapenemase-encoding genes.37,38 In fact, the genome of S. maltophilia encodes B Zn2+-dependent metalloenzyme that hydrolyzes all classes of β-lactams except the monobactams.39 Besides, a remarkably high level of resistance of the clinical isolates to amoxycillin/clavulanic acid and trimethoprim/sulfamethoxazole, which are primarily responsible for the development of community-acquired pneumonia, was reported in Ethiopia.8 Hence, our results were consistent with studies conducted in Ethiopia.
Among the predominant isolates, K. pneumoniae and S. marcescens showed complete resistance to tetracycline (100%). Additionally, tetracycline and chloramphenicol resistant S. maltophilia might impose a further risk to the treatment of immunocompromised patients. Although carbapenems are considered the most effective drugs for treating A. pittii, imipenem-resistant A. pittii leaves limited therapeutic options.40 Thus, the spread of MDR A. pittii species in Northern Gondar could lead to more infections in patients with underlying health problems. Finally, K. cryocrescens, which is considered to be a very rare pathogen, also showed resistance to several drugs, including gentamycin and trimethoprim/sulfamethoxazole, although its frequency of isolation is low.
Conclusion
The findings revealed that the predominant imipenem-resistant bacteria recovered from sputum samples were A. baumannii, S. marcescens, K. pneumoniae, and P. aeruginosa. Besides imipenem, resistance to ampicillin, amoxycillin/clavulanic acid, and trimethoprim/sulfamethoxazole was also high. Remarkably, many bacterial isolates showed susceptibility to gentamycin. Hence, local empirical prescriptions should correspond to the results of antibiotic sensitivity to prevent the development of antibiotic resistance in the region and to reduce the nosocomial and community-acquired infection caused by the identified bacteria. It is also necessary to continuously monitor antibiotic resistance in the region by implementing effective antimicrobial stewardship programs. Future work will identify the genetic diversity and clonality of specific imipenem-resistant bacterial species from different regions of Ethiopia using whole-genome sequencing.
Abbreviations
ATCC, American Type Culture Collection; AMR, antimicrobial resistance; CLSI, Clinical Laboratory Standard Institute; h, hours; LB, Luria-Bertani media; min, minutes; MDR, multidrug resistance; PCR, polymerase chain reaction; TSI, triple sugar iron agar.
Ethics Approval and Consent to Participate
Done as described earlier.
Acknowledgments
The authors would like to thank Prof. Wolfgang Streit from the University of Hamburg for unreserved support with the 16S rRNA sequencing. We also thank the laboratory staff of the two health centers for their support in specimen collection.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Author Details
Addis Ababa Science and Technology University, Department of Biotechnology, P.O.Box.16417, Addis Ababa, Ethiopia, and University of Gondar, Department of Biology, P.O.Box.196, Gondar, Ethiopia
Funding
This study was partially funded by the University of Gondar.
Disclosure
The authors declare that they have no competing interests.
References
1. Cassini A, Högberg LD, Plachouras D, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66. doi:10.1016/S1473-3099(18)30605-4
2. U.S. Centers for Disease Control and Prevention. About Antimicrobial Resistance - Biggest Threats and Data 2019 AR Threats Report. Centers Dis Control Prev. 2019:125.
3. World Bank Group. Drug-Resistant Infections: A Threat to Our Economic Future. World Bank Report.; 2017.
4. Founou RC, Founou LL, Essack SY, Butaye P. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS One. 2017;12(12):12. doi:10.1371/journal.pone.0189621
5. Guitor AK, Wright GD. Antimicrobial Resistance and Respiratory Infections. Chest. 2018;154(5):1202–1212. doi:10.1016/j.chest.2018.06.019
6. Ahmed SMA-Z, Abdelrahman SS, Saad DM, Osman IS, Osman MG, Khalil EAG. Etiological Trends and Patterns of Antimicrobial Resistance in Respiratory Infections. Open Microbiol J. 2018;12(1):34–40. doi:10.2174/1874285801812010034
7. Beyene G, Regasa B, Yilma D, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from community-acquired pneumonia patients in Jimma University Specialized Hospital, Jimma, Ethiopia. Saudi J Health Sci. 2015;4(1):59. doi:10.4103/2278-0521.151411
8. Temesgen D, Bereded F, Derbie A, Biadglegne F. Bacteriology of community acquired pneumonia in adult patients at Felege Hiwot Referral Hospital, Northwest Ethiopia: A cross-sectional study. Antimicrob Resist Infect Control. 2019;8(1):101. doi:10.1186/s13756-019-0560-0
9. Murray BE, Wood AJJ. Vancomycin-resistant enterococcal infections. N Engl J Med. 2000;342(10):710–721. doi:10.1056/NEJM200003093421007
10. Fink MP, Snydman DR, Niederman MS, et al. Treatment of severe pneumonia in hospitalized patients: results of a multicenter, randomized, double-blind trial comparing intravenous ciprofloxacin with imipenem-cilastatin. The Severe Pneumonia Study Group. Antimicrob Agents Chemother. 1994;38(3):547–557. doi:10.1128/AAC.38.3.547
11. Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6:47.
12. Singh N, Manchanda V. Control of multidrug-resistant Gram-negative bacteria in low- and middle-income countries—high impact interventions without much resources. Clin Microbiol Infect. 2017;23(4):216–218. doi:10.1016/j.cmi.2017.02.034
13. Humphries RM, Ambler J, Mitchell SL, et al. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol. 2018;56(4):e01934–17.
14. Odds F. Biochemical Tests for Identification of Medical Bacteria. J Clin Pathol. 1981;34(5):572. doi:10.1136/jcp.34.5.572-a
15. Amoli RI, Nowroozi J, Sabokbar A, Rajabniya R. Isolation of Stenotrophomonas maltophilia from clinical samples: an investigation of patterns motility and production of melanin pigment. Asian Pac J Trop Biomed. 2017;7(9):826–830. doi:10.1016/j.apjtb.2017.08.012
16. Thomas R, Hamat RA, Neela V. Extracellular enzyme profiling of Stenotrophomonas maltophilia clinical isolates. Virulence. 2014;5(2):326–330. doi:10.4161/viru.27724
17. Welker E, Domfeh Y, Tyagi D, Sinha S, Fisher N. Genetic Manipulation of Stenotrophomonas maltophilia. Curr Protoc Microbiol. 2015;37(1):
18. Juretschko S, Timmermann G, Schmid M, et al. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64(8):3042–3051. doi:10.1128/AEM.64.8.3042-3051.1998
19. Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients.. Appl Environ Microbiol. 1997;63(8):3233–3241. doi:10.1128/AEM.63.8.3233-3241.1997
20. Allers E, Niesner C, Wild C, Pernthaler J. Microbes enriched in seawater after addition of coral mucus. Appl Environ Microbiol. 2008;74(10):3274–3278. doi:10.1128/AEM.01870-07
21. Pompilio A, Crocetta V, De Nicola S, Verginelli F, Fiscarelli E, Di Bonaventura G. Cooperative pathogenicity in cystic fibrosis: Stenotrophomonas maltophilia modulates Pseudomonas aeruginosa virulence in mixed biofilm. Front Microbiol. 2015;6:951.
22. Azene MK, Beyene BA. Bacteriology and antibiogram of pathogens from wound infections at Dessie Laboratory, North East Ethiopia. Tanzan J Health Res. 2011;13(4):68–74. doi:10.4314/thrb.v13i4.64901
23. Mohammed A, Seid ME, Gebrecherkos T, Tiruneh M, Moges F. Bacterial Isolates and Their Antimicrobial Susceptibility Patterns of Wound Infections among Inpatients and Outpatients Attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int J Microbiol. 2017;2017:8953829. doi:10.1155/2017/8953829
24. Muluye D, Wondimeneh Y, Ferede G, et al. Bacterial isolates and their antibiotic susceptibility patterns among patients with pus and/or wound discharge at Gondar University hospital. BMC Res Notes. 2014;7(1):619. doi:10.1186/1756-0500-7-619
25. Nathan AM, Teh CSJ, Jabar KA, et al. Bacterial pneumonia and its associated factors in children from a developing country: A prospective cohort study. PLoS One. 2020;15(2):e0228056. doi:10.1371/journal.pone.0228056
26. Seboxa T, Amogne W, Abebe W, et al. High mortality from blood stream infection in Addis Ababa, Ethiopia, is due to antimicrobial resistance. PLoS One. 2015;10(12):e0144944. doi:10.1371/journal.pone.0144944
27. Troillet N, Samore MH, Carmeli CY. Imipenem-Resistant Pseudomonas Risk Factors and Antibiotic Susceptibility Patterns. Clin Infect Dis. 1997;25(5):1094–1098. doi:10.1086/516092
28. Meyer E, Schwab F, Schroeren-Boersch B, Gastmeier P. Dramatic increase of third-generation cephalosporin-resistant E. coli in German intensive care units: secular trends in antibiotic drug use and bacterial resistance, 2001 to 2008. Crit Care. 2010;14(3):R113. doi:10.1186/cc9062
29. Armand-Lefèvre L, Angebault C, Barbier F, et al. Emergence of imipenem-resistant Gram-negative bacilli in intestinal flora of intensive care patients. Antimicrob Agents Chemother. 2013;57(3):1488–1495. doi:10.1128/AAC.01823-12
30. Ssekatawa K, Byarugaba DK, Wampande E, Ejobi F. A systematic review: the current status of carbapenem resistance in East Africa. BMC Res Notes. 2018;11(1):629. doi:10.1186/s13104-018-3738-2
31. Agyepong N, Govinden U, Owusu-Ofori A, Essack SY. Multidrug-resistant Gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrob Resist Infect Control. 2018;7(1):37. doi:10.1186/s13756-018-0324-2
32. Legese MH, Weldearegay GM, Asrat D Extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae among Ethiopian children. Infect Drug Resist. 2017;10:27–34. doi:10.2147/IDR.S127177
33. Codjoe F, Donkor E. Carbapenem Resistance: A Review. Med Sci. 2017;6(1):1.
34. Khorshidi A, Sharif AR. Imipenem resistance among Gram-negative and Gram-positive bacteria in hospitalized patients. Iran J Public Health. 2010;39(2):110–113.
35. Woodford N, Xu-McCrae L, Mushtaq S, et al. Prevalence of carbapenem resistance and carbapenemase production among Enterobacteriaceae isolated from urine in the UK: results of the UK infection-Carbapenem Resistance Evaluation Surveillance Trial (iCREST-UK). J Antimicrob Chemother. 2018;73(3):698–702. doi:10.1093/jac/dkx471
36. Beyene D, Bitew A, Fantew S, Mihret A, Evans M, Ruiz-Rodriguez M. Multidrug-resistant profile and prevalence of extended spectrum β-lactamase and carbapenemase production in fermentative Gram-negative bacilli recovered from patients and specimens referred to National Reference Laboratory, Addis Ababa, Ethiopia. PLoS One. 2019;14(9):e0222911. doi:10.1371/journal.pone.0222911
37. Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3(1):15–21.
38. Tängdén T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med. 2015;277(5):501–512. doi:10.1111/joim.12342
39. Crossman LC, Gould VC, Dow JM, et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008;9(4):R74. doi:10.1186/gb-2008-9-4-r74
40. Chusri S, Chongsuvivatwong V, Rivera JI, et al. Clinical outcomes of hospital-acquired infection with Acinetobacter nosocomialis and Acinetobacter pittii. Antimicrob Agents Chemother. 2014;58(7):4172–4179. doi:10.1128/AAC.02992-14
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

