Back to Journals » Infection and Drug Resistance » Volume 12
Evaluating the antimicrobial resistance patterns among major bacterial pathogens isolated from clinical specimens taken from patients in Mofid Children’s Hospital, Tehran, Iran: 2013–2018
Authors Azimi T , Maham S, Fallah F, Azimi L , Gholinejad Z
Received 10 May 2019
Accepted for publication 21 June 2019
Published 17 July 2019 Volume 2019:12 Pages 2089—2102
DOI https://doi.org/10.2147/IDR.S215329
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Taher Azimi,1,2 Saied Maham,1 Fatemeh Fallah,1 Leila Azimi,1 Zari Gholinejad1
1Pediatric Infections Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran; 2Department of Pathobiology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran
Background: This study evaluates the epidemiology and antimicrobial resistance profile of Gram-negative bacteria (GNB) and Gram-positive bacteria (GPB) isolated from clinical specimens in children admitted to Mofid Children’s Hospital.
Methods: This was a retrospective study of the patients’ clinical specimens collected from January 2013 until the end of December 2018. All specimens were evaluated to determine the presence of infection-causing agents using a BACTEC 9120 blood culture. Isolation and identification of bacterial strains were performed using conventional biochemical tests. Antibiotic resistance was determined using Kirby–Bauer disk diffusion and broth microdilution methods. Results were interpreted according to CLSI and EUCAST.
Results: A total of 1130 different pathogenic bacteria were detected from 14,690 different clinical specimens and the overall detection rate was 7.7% (1130/14,690). Among bacterial pathogen isolated from clinical specimens, 55% (n=622) were GNB and 45% (n=508) were GPB. The predominant GNB isolates were Pseudomonas aeruginosa, Klebsiella spp., Acinetobacter baumannii, Escherichia coli, Enterobacter spp., Citrobacter spp., respectively. Among GPB, CoNS was the most frequent and Enterococcus spp. was found to have low levels of resistance to linezolid. In GNB, most A. baumannii and P. aeruginosa were ceftriaxone resistant. P. aeruginosa was found to have low levels of resistance to levofloxacin and ciprofloxacin.
Conclusions: Our findings revealed that the resistance rate among GNB and GPB associated with different infections in children is very high. These results suggest a constant screening and follow-up programs for the detection of antibiotic resistance, and it also suggests to develop antimicrobial stewardship programs in Tehran, Iran.
Keywords: Gram-negative bacteria, Gram-positive , bacteria, bacterial, infection, antimicrobial, resistance, children, Iran
Introduction
Nowadays, antimicrobial resistance (AMR) is considered as a main public health threat,1–5 also AMR bacteria in different hospital wards are increasing significantly.6–8 Based on a published study, 700,000 deaths are reported annually due to AMR, and it has been predicted that if appropriate control and prevention measures are not taken, AMR would become one of the main reasons of death among hospitalized or non-hospitalized patients in developing and developed countries.9 Proper antibiotic usage and administration are essential for treatment of bacterial infections.10,11 Thus, inappropriate prescription and misuse of antibiotics could contribute to the emergence of AMR pathogenic bacteria, restriction of therapeutic options, increase of hospitalization time and high treatment costs and finally a greater death rate.12–14 Bacteria are one of the main causes of infections in humans.15–17 Children are usually considered as the most vulnerable group to bacterial infections.12,18 According to the global action plan on AMR endorsed by WHO, it is important to raise awareness on AMR through monitoring and research programs in different parts of the world.19–21 AMR monitoring is critical and has several benefits including: 1) providing data on bacterial resistance rate, 2) helping select appropriate antibiotics and subsequently reduce AMR rate,22 3) reduction in hospitalization rate and treatment costs, and 4) decrease in death rate.1,11 Therefore, the current study evaluates the epidemiology and AMR profile of the main pathogenic bacteria isolated from hospitalized children from January 2013 until the end of December 2018 in Mofid Children’s Hospital, Tehran Iran.
Materials and methods
Study design and identification of microorganisms
This was a retrospective study of the patients’ clinical specimens collected over a 6-year period from January 2013 until the end of December 2018 in Mofid children’s hospital. This research used microbiological lab data of 14,690 various clinical specimens collected from different hospital wards in Mofid children’s hospital in Tehran. Clinical specimens were collected in BACTEC standard culture vials and were incubated at 37°C in BACTEC automated system. Moreover, BACTEC system database was studied and after patients with positive samples were detected, some information related to these patients suchas sex, age, type of specimens and their hospitalized ward was obtained. In the next step, the positive samples were sub-cultured on specific medium including MacConkey agar, chocolate agar, mannitol salt agar and blood agar plates, and then Gram staining of bacterial colonies were performed. Isolation and identification of different bacterial strains of positive cultures were performed using conventional biochemical tests including IMVIC (Indole, Methyl red, Voges proskauer and Citrate) test, catalase and oxidase test, growth on Triple Sugar Iron Agar and Kligler Iron Agar, Bile esculin agar, SH2 production, motility test, growth on 6% NaCl and DNase test.
Antibiotic susceptibility testing
The antibiotic resistance of the isolates was determined using Kirby–Bauer disk diffusion method (DDM) and broth microdilution method (for evaluating colistin and PB susceptibility in Gram-negative bacteria (GNB)); the results of DDM method were then interpreted according to Clinical and Laboratory Standards Institute (CLSI) criteria.
Moreover, interpretation of colistin MIC results was performed according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (susceptible, ≤2 mg/L; resistant, >2 mg/L). On the other hand, considering that CLSI and EUCAST do not have interpretive criteria for polymyxin B for Enterobacteriaceae, a breakpoint of >2 mg/L for resistant isolates and ≤2 mg/L for susceptible isolates were used. The antibiotic discs and powders were purchased from MAST Company and Sigma (Sigma–Aldrich, cat No. PZ0021). The Gram-positive and -negative bacterial isolates including Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853) and Staphylococcus aureus subsp. aureus ATCC 25923 were used as quality control for DDM and MIC tests. Due to annual changes in hospital policies, the applied antibiotics for treatment of various bacterial infections varied too.
The antimicrobial susceptibility for GNB and Gram-positive bacteria (GPB) was determined using the following antibiotic disks: ciprofloxacin (CIP); piperacillin/tazobactam (PTZ); tetracycline (TET); chloramphenicol (CHL); amikacin (AK); cefotaxime (CTX); ceftazidime (CAZ); cefepime (CPM); levofloxacin (LEV); trimethoprim-sulfamethoxazole (SXT); tobramycin (TOB); doxycycline (DOX); meropenem (MRP); doripenem (DOR); imipenem (IPM); ampicillin-sulbactam (SAM); ceftriaxone (CRO); cefuroxime (CXM); ampicillin (AMP); amoxicillin-clavulanic acid (AUG); nitrofurantoin (NI); gentamicin (GM); nalidixic acid (NA); cefazolin (CZ); azithromycin (AZM); cefoxitin (FOX); oxacillin (OX); erythromycin (ERY); quinupristin-dalfopristin (RP); vancomycin (VA); cefdinir (CD); clindamycin (CLI); ofloxacin (OFX); linezolid (LZD); penicillin (PEN); clarithromycin (CLa); cefpodoxime (CPd); norfloxacin (NOR); ticarcillin-clavulanic acid (TIM); aztreonam (ATM). The results of the research were documented as either susceptible (S), intermediate (I) or resistant (R). According to the European Centre for Disease Prevention and Control (ECDC) and the US Centers for Disease Control and Prevention (CDC), the identification of multidrug-resistant (MDR) isolates was conducted and GNB were selected as MDR, which were resistant to at least one antimicrobial among at least three or more drug categories.
Statistical analysis
The patients' information such as gender, age, type of specimens, the hospitalized ward and antibiotic susceptibility profiles were all collected from the hospital database and were analyzed using the statistical package SPSS v.23.0 (SPSS Inc., Chicago, IL, USA).
Results
Number and distribution of specimens and positive cultures
During this 6-year period, a total of 14,690 different clinical cultures were collected from January 2013 until the end of December 2018. Among which, 1130 (7.7%) cultures were positive from which various bacteria were isolated. Among GPB, about 58.8% and 41.2% of the total positive cultures were from male and female samples, respectively. On the other hand, among GNB, approximately 55% and 45% of the total positive cultures belonged to male and female samples, respectively (Table 2). In positive cases, the mean age was 11.2 years old (1 month to 15 years old). Various bacteria recovered from different hospital wards are: Hematology (n=151; 13.3%), Emergency (n=43; 3.8%), Gastroenterology (n=139; 12.3%), ICU (n=22; 2%), Infectious (n=287; 25.3%), Nephrology (n=33; 3%), Neurology (n=35; 3%), NICU (n=36; 3.1%), Renal unit (n=14; 1.2%), PICU (n=242; 21.4%), Rheumatology (n=15; 1.3%), Surgery (n=97; 8.5%), Oncology (n=10; 0.8%) and Urology (n=2; 0.17%). The frequency of various clinical samples isolated from bacterial strains was as follows: blood (n=1003; 88.7%), cerebrospinal fluid (CSF) (n=61; 5.3%), pleural fluid (n=46; 4%), dialysis fluid (n=1; 0.09%), luminal sample (n=8; 0.7%) and shunt (n=11; 1%). The frequency of GNB and GPB in various positive clinical samples and different hospital wards are shown in Table 2. Evaluation of the distribution of GPB and GNB isolates in different hospital wards has shown that most clinical isolates (25.9% and 24.7%, respectively) were identified in the infection ward.
 |
Table 1 The characteristic of clinical specimens and isolated bacteria in 2013–2018 |
 |  |  |
Table 2 Antimicrobial resistance of isolated GPB from clinical specimens in 2013–2018 |
Pathogen distribution
GNB and GPB comprised 55% (n=622) and 45% (n=508) of the total bacteria, respectively. The isolated GNB included P. aeruginosa (n=282; 45.3%), Klebsiella spp. (n=100; 16.07%), Acinetobacter baumannii (n=83; 13.3%), E. coli (n=59; 9.4%), Enterobacter spp. (n=45; 7.2%), Citrobacter spp. (n=28; 4.5%), Burkholderia spp. (n=20; 3.2%) and Proteus spp. (n=5; 0.8%). Moreover, the most predominant isolated GPB was coagulase-negative Staphylococcus (CoNS) (n=368; 72.4%), Enterococcus spp. (n=71; 13.9%) and S. aureus (n=69; 13.5%). The highest number (n=311) of strains was isolated in 2016 and the lowest (n=85) in 2013. Moreover among GNB and GPB, P. aeruginosa and CoNS were the most frequent pathogens, respectively (Table 1).
Antimicrobial susceptibility
Resistance rates of GPB to antimicrobials
The resistance rates of the isolated GPB to commonly used antimicrobials are shown in Table 2. In S. aureus, isolated from different specimens, the highest resistance rates belonged to oxacillin (n=43/64; 67.2%). However, S. aureus was found to have low levels of resistance to vancomycin (n=1/67; 1.5%) and linezolid (n=3/66; 4.5%). In addition, CoNS strains showed a high level of resistance to oxacillin (n=273/318; 85.8%) and ampicillin (n=37/46; 80.4%). vancomycin (n=13/360; 3.6%) and linezolid (n=15/322; 4.7%) were the most effective antimicrobial agents on CoNS.
Enterococcus spp. was 100% resistant to ofloxacin (n=11/11;1 00%), clarithromycin (n=8/8; 100%), cefotaxime (n=3/3;100%), nitrofurantoin (n=11/11; 100%), amikacin (n=11/11; 100%), azithromycin (n=5/5; 100%), clindamycin (n=3/3; 100%), piperacillin/tazobactam (n=2/2; 100%), and doxycycline (n=1/1; 100%) which showed high levels of resistance to trimethoprim-sulfamethoxazole (n=13/14; 92.9%) and quinupristin-dalfopristin (n=10/11; 90.9%). However, Enterococcus spp. was found to have low levels of resistance (n=4/59; 6.8%) to linezolid.
Resistance rates of GNB to antimicrobials
Overall, among GNB, P. aeruginosa was 100% resistant to amoxicillin-clavulanic acid (n=26/26; 100%) and ticarcillin-clavulanic acid (n=17/17; 100%) and showed a high level of resistance to cefazolin (n=45/46; 97.8%) and ceftriaxone (n=29/31; 93.5%). P. aeruginosa was found to have low levels of resistance to levofloxacin (n=20/154; 13%) and ciprofloxacin (n=43/257; 16.7%), respectively. However, 29.8% (n=84/282) of P. aeruginosa was MDR.
Levofloxacin (n=19/49; 38.8%) was the most effective antimicrobial agents on A. baumannii. However, the resistance level to cefuroxime (n=26/26; 100%), cefotaxime (n=35/36; 97.2%) and meropenem (n=64/68; 94.1%) was high. Moreover, 62.7% (n=52/83) of A. baumannii was MDR.
The resistance rates of Klebsiella spp. to levofloxacin, imipenem and ciprofloxacin were 12.8%, 18.5% and 21%, respectively. In addition, a high level of resistance to cefpodoxime (n=6/6; 100%), ampicillin (n=61/65; 93.8%) and cefazolin (n=39/44; 88.6%) was detected; 27% (n=27/100) of Klebsiella spp. strains were MDR.
All of the tested isolates of Enterobacter spp. showed resistance to ampicillin (n=23/23; 100%) and tetracycline (n=14/14; 100%) and showed a high level of resistance to ampicillin-sulbactam (n=29/31; 93.5%). Levofloxacin (n=2/12; 16.7%), imipenem (n=7/38; 18.4%) and nalidixic acid (n=1/5; 20%) were the most effective antimicrobial agents on Enterobacter spp. Moreover, 26.7% (n=12/45) of Enterobacter spp. were MDR.
Proteus spp. was found to have low levels of resistance to imipenem (n=0/5; 0%), ciprofloxacin (n=0/3; 0%), amikacin (n=0/2; 0%) and levofloxacin (n=0/2; 0%); and 20% (n=1/5) of Proteus spp. strains were MDR.
For E. coli, the resistance rate was 94.7% (n=18/19) for trimethoprim sulfamethoxazole; 94.6% (n=35/37) for ampicillin; 92% (n=23/25) for amoxicillin-clavulanic acid and >80% for several antibiotics such as ceftriaxone, ampicillin-sulbactam, cefuroxime and aztreonam. A high proportion of E. coli strains (n=30/59; 50.8%) was MDR.
The resistance rates of Burkholderia spp. to trimethoprim-sulfamethoxazole, cefotaxime and ticarcillin-clavulanic acid were 100%, 88.9% and 87.5%, respectively. Ciprofloxacin (n=4/20; 20%), levofloxacin (n=3/12; 25%) and colistin (n=4/15; 26.7%) were the most active antibiotics against Burkholderia spp, and 30% (n=6/20) were MDR.
Ciprofloxacin and levofloxacin were the most active antibiotic against Citrobacter spp. The resistance level of this organism to ampicillin (n=24/24) and cefazolin (n=7/7) was 100%, and 64.3% (n=18/28). The resistance rates of each GNB to commonly used antimicrobials are shown in Table 3.
 |
Table 3 Antimicrobial resistance of isolated GNB from clinical specimens in 2013–2018 |
Time trends in antibiotic resistance among GPB and GNB isolated from Clinical specimens in 2013–2018
The time trend analysis of antibiotic resistance among GPB and GNB is shown in Tables 4 and 5, respectively. Among GPB, results show that S. aureus isolates had the highest resistance rate to linezolid (R=50%) in 2017. Moreover, vancomycin is an effective antibiotics against GPB and has a lowest resistance rate (R=4.5% in 2017) against S. aureus isolates.
 |
Table 4 Trends in antibiotic resistance among GPB isolated from clinical specimens in 2013–2018 |
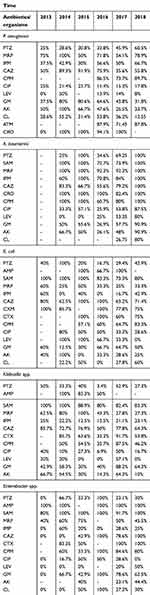 |
Table 5 Trends in antibiotic resistance among GNB isolated from clinical specimens in 2013–2018 |
In 2015, Enterococcus isolates had the highest resistance to vancomycin (R=63.6%), and levofloxacin was the most effective antibiotic against Enterococcus isolates in 2017 (resistance has not been seen). In CoNS isolates, the highest and lowest resistance to vancomycin was seen in 2013 and 2018, respectively. Linezolid was an effective antibiotic against CoNS isolates in 2013 (resistance has not been seen) and the highest resistance was seen (R=15.4%) in 2014. Among GNB, levofloxacin was the most effective antibiotic against P. aeruginosa isolates in 2013 and resistance has not been seen. However, in 2014 levofloxacin has the highest resistance (R=50%) against this pathogen. In P. aeruginosa isolates, ciprofloxacin has the highest and lowest resistance in 2013 and 2016, respectively. Our analysis showed that A. baumannii has the highest resistance to levofloxacin (R=80%) in 2018.
Discussion
This study examined the prevalence of antibiotic resistance among main pathogenic bacteria isolated from hospitalized children in Mofid Children’s Hospital, Tehran, Iran. Given that these antibiotic resistance for GNB and GPB can cause severe infections in hospitalized patients, especially in neonates and children, the presence and distribution of these agents is one of the main concerns for physicians.23,24 Since the application of several categories of antibiotics is not permissible in neonates and children and since there are different patterns of AMR in various areas, selecting and prescribing appropriate antibiotics for the treatment of various infections in pediatric patients is challenging.15,25 Moreover, knowing AMR patterns can help physicians and policy makers to find solutions for resistance problems in their countries.23,26 Lack of general AMR surveillance programs in developing and several developed countries will lead to inappropriate use among patients and health care staff.27 Therefore, investigating AMR patterns are very critical and important, mainly in developing countries such as Iran, where there is no systematic guidelines for antibiotic usage. On the other hand, it is necessary to analyze the patterns of antibiotic resistance for GPB and GNB at Mofid Children’s Hospital of Tehran, capital of Iran, during 2013–2018, which can be a valuable model for both clinicians and policy makers in implementing empirical therapy. The result of our study revealed that among 14,690 different clinical samples from unique patients, 1130 (7.7%) cultures were positive from which various bacteria were isolated. The low rate of positive culture in the current study can be due to several reasons: 1) our study used various types of clinical samples such as blood, CSF, pleural fluid, dialysis fluid and luminal fluid in which the rate of pathogens in these specimens is different, 2) effective training for the correct administration of antibiotic, 3) better management and control of infection, and 4) pre-hospitalization use of antibiotics. The amount of GNB and GPB isolates among all positive cultures were 55% (n=622) and 45% (n=508), respectively, and it was detected that GNBs are frequently isolated in positive cultures. In the present study, P. aeruginosa and CoNS were the most frequent pathogens among GNB and GPB, respectively, which is in agreement with a study conducted by Mahmoudi et al (2011–2016 Tehran, Iran).12 However, in the investigations carried out by Ebrahim-Saraie et al and Alam et al, Acinetobacter spp. was the most common GNB in positive culture specimens.28,29 The result of a published study revealed that E. coli was the most frequent Gram-negative pathogen in positive cultures of the specimens30. The detected differences in proportions of GNB and GPB could be due to the diversity of specimen type, specimen size and applied detecting methods. Among different antibiotics that were tested against P. aeruginosa, levofloxacin and ciprofloxacin were the effective antibiotics, respectively. On the other hand, P. aeruginosa showed a high level of resistance to ticarcillin-clavulanic acid, cefazolin and ceftriaxone. Similarly, A. baumannii showed the lowest and highest resistance rate to levofloxacin and meropenem, respectively.
Results of time trend analyses showed that levofloxacin resistance rate against P. aeruginosa has decreased from 2014 to 2018. Moreover, these results showed that levofloxacin resistance rate against A. baumannii has increased from 2016 to 2018.
According to this date, suitable antibiotic selection is significant and vital in the treatment of bacterial infections. Therefore, awareness regarding antibiotic resistance patterns in pathogenic bacteria can be helpful in making the right therapeutic choice. The results have also shown that probably CoNS isolated from clinical specimens was considered as a common contaminant. Therefore, more effective measures such as hand hygiene of health care workers, regular disinfection of medical devices, and disinfection of sampling site need to be taken during sampling. However, albeit rare, CoNS can cause several infections including skin and soft tissue infections and thus should not at all times be considered as contaminants.31 Persistent CoNS infection is probably related to various severe complications such as embolic complications, metastatic seeding and septic thrombophlebitis.32 Therefore, the evaluation of CoNS medical correlation is a challenging problem. In medical diagnostic laboratories, the main diagnostic challenge is to evaluate whether an expected CoNS isolate represents: 1) a common colonization of the skin, soft tissue or mucous membranes, 2) a contamination of the specimen throughout sample collection, handling and processing, or 3) clinically important infection.33 In the case of coinfection of CoNS with other bacterial infections (polymicrobial infections by CoNS), different bacteria isolates showed various susceptibility and resistance patterns, this diagnostic challenging situation becomes even extra intricate.32,33 A close collaboration between medicians and diagnostic laboratory specialists can solve this medical and diagnostic problem. In case of false-positive CoNS cases, the patients are treated with several antibiotics, it is predicted that besides additional costs, excessive antibiotic selection pressures occur which can lead to the emergence of antibiotic resistance.34 Therefore, it is important to answer the question that CoNS isolated from a clinical specimen is a real infection or only a common contamination or skin colonization. Some of the main factors useful in the prediction of real infection are: 1) similar strains being isolated repeatedly during course of an infection after the isolation of a strain in pure culture from the infected site, 2) in bloodstream infections, patients should have clinical evidence of the infection with one positive blood culture or only two positive CoNS blood cultures within 5 days, and 3) if CoNS is isolated from a skin or soft tissue bacterial culture of a suspected infectious lesion, the isolated organism should be proposed as a pathogen and suitable treatment should be initiated.35–37
Among different-tested antibiotics, the results of our study have revealed that linezolid and vancomycin are effective antibiotics against S. aureus and Enterococcus spp, which was in agreement with the rates reported by Dharmapalan et al from India,38 He et al from China,39 Lei Tian et al from China,40 and Al-Naqshbandi et al from Iraq.41 However, the results of several studies were not consistent with our research and it has been reported that the resistance to vancomycin is high.42,43 In the present study, one case of vancomycin resistant to S. aureus (VRSA) was observed; however, vancomycin resistance in Enterococcus spp. was much higher (37/69); 53.6% of Enterococcus spp. isolates were vancomycin-resistant. Although the identification of Enterococcus spp. was not performed to species level, we proposed that most vancomycin-resistant isolates are more likely to be Enterococcus faecium. According to several published studies and reports, effective measures were taken to decrease the risk of VRSA in several countries such as the USA, also some guidelines were developed to control the infections caused by these pathogenic microorganisms.15 Thus, we suggest similar guidelines and programs designed for children and neonates patients in Tehran, Iran. All in all, the results of the current study revealed that ticarcillin-clavulanic acid, ampicillin, amoxicillin-clavulanic acid, cefazolin and ceftriaxone are ineffective antibiotics against GNB. Notably, in different hospitals in Tehran, these antibiotics are frequently used to control various infections especially sepsis and septicemia. It is well understood that resistance to these antibiotics increases daily, and it is the consequence of selective pressure excreted via bystander selection and abuse or overuse of antibiotics.44 According to high antibiotic resistance among bacteria, in order to prevent undesirable effects of sepsis and septicemia, as well as in order to reduce the mortality rate due to these infections, precise detection and use of effective antibiotics for an efficient treatment are critical.8,45–47 Consequently, awareness of the antibiotic resistance patterns among common pathogens, holding workshops to correct prescription of empiric treatment, and changes in antimicrobial use are warranted and highly recommended. Finally, the results of DDM and MIC tests are of great importance, and individuals' free access to antibiotics should be prevented. In this study, we have revealed that GNB and GPB are resistant to various groups of antibiotics; however, it should be noted that these bacteria have two types of antibiotic resistance: acquired resistance and intrinsic resistance. For instance, according to EUCAST guideline, most GNB (Enterobacteriaceae, Pseudomonas spp.) have an intrinsic resistance to various antibiotics including penicillin G, oxacillin, macrolides (eg, azithromycin, erythromycin, tylosin), lincosamides (eg, lincomycin, clindamycin), streptogramins (eg, virginiamycin), glycopeptides (eg, vancomycin) and bacitracin. Moreover, based on these guidelines, most GPB are intrinsically resistant to polymyxins and quinolones/fluoroquinolones (eg, enrofloxacin, ciprofloxacin, difloxacin, marbofloxacin).48 Therefore, these resistances should be known by clinicians in order to avoid unsuitable and ineffective therapy.
Our study also revealed that colistin, in comparison with levofloxacin and ciprofloxacin, has the highest resistance rate. These finding were in contrast with the results of Mahmoudi et al from Iran12 and Dharmapalan et al from India.38 In the current study, among GNB, Proteus spp. and Citrobacter spp. have the lowest and highest rate of MDR, respectively. The infection with MDR bacteria has several unfortunate consequences such as increased hospitalization, increase in health care and hospitalization costs, reduction in success rate of the infection treatments and increase in morbidity and mortality rates.12 Moreover, according to results obtained from the prevalence of MDR bacteria that revealed the incidence of MDR bacteria is increasing and by considering the fact that there are limited therapeutic options for MDR bacterial infections, serious measures such as well-controlled clinical trials of combinations of existing antibiotics (for instance colistin plus rifampicin against MDR P. aeruginosa infections, colistin plus vancomycin, colistin–carbapenem, trimethoprim-sulfamethoxazole with colistin or fosfomycin and aminoglycoside against enterobacteriaceae infection or combination of cloxacillin (150 mg/kg/day dose), along with gentamicin in CoNS blood infection should be urgently taken.49 On the other hand, systematic surveillance of hospitals and community-acquired infections, hospital waste management, monitoring the use of antibiotics, monitoring and evaluating antibiotic sensitivity, and preparing reliable antibiotic strategies are necessary.
Our study has several limitations including: 1) it was not a cross-sectional investigation and had a retrospective nature; therefore, given the incompleteness of databases, several main clinical data of patients including results of treatment and mortality rate due to GPB and GNB were not available and we could not include this information in our research, 2) there was no control group for some variables in this study, 3) it also lacked access to patients information such as treatment outcomes and mortality rate due to GNB and GPB, 4) and finally the classification of GNB and GPB, because of their unknown origins (hospital or community-acquired infections) was not performed, 5) the identification of Citrobacter, Enterococcus, CoNS, Enterobacter, Klebsiella, Burkholderia and Proteus were not performed to species level; therefore, determining the infection rates and antibiotic resistance patterns of different species of these bacteria was impossible. 6) Considering several factors including annual changes in hospital policies, availability of antibiotics in laboratory and based on the antibiotic resistance patterns reported by laboratory experts as well as physicians’ recommendations, the utilized antimicrobial agents and the number of tested isolates varied from year to year. Consequently, in the case of some antibiotics including imipenem, ceftriaxone, and several other antibiotics, there were few-studied cases; so, their susceptibility and resistance patterns do not absolutely reflect their situations. Therefore, the applied antibiotics for the treatment of various bacterial infection varied too. There have been few studies on several antibiotics including cefotaxime, ceftriaxone, clarithromycin and nitrofurantoin among GPB, and polymyxin B, cefpodoxime and azithromycin among GNB, and thus their AMR patterns do not fully reflect their resistance or susceptibility positions. 7) Moreover, based on the fact that we did not have full access to patients information such as treatment outcomes, mortality rate, etc., no specific analysis was performed.
Acknowledgments
We would like to thank “Pediatric Infections Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran and Division of Microbiology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran” for their kind cooperation. The authors received no specific funding for this work.
Author contributions
All authors contributed to data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work. Taher Azimi and Saied Maham share first authorship.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gandra S, Mojica N, Klein EY, et al. Trends in antibiotic resistance among major bacterial pathogens isolated from blood cultures tested at a large private laboratory network in India, 2008–2014. Int J Infect Dis. 2016;50:75–82. doi:10.1016/j.ijid.2016.08.002
2. O’Brien TF, Clark A, Peters R, Stelling J. Why surveillance of antimicrobial resistance needs to be automated and comprehensive. J Glob Antimicrob Resist. 2018;17:8–15.
3. Pormohammad A, Nasiri MJ, Azimi T. Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: a systematic review and meta-analysis. Infect Drug Resist. 2019;12:1181. doi:10.2147/IDR.S201324
4. Sharahi JY, Azimi T, Shariati A, Safari H, Tehrani MK, Hashemi A. Advanced strategies for combating bacterial biofilms. J Cell Physiol. 2019;234:14689–14708. doi:10.1002/jcp.v234.9
5. Azimi T, Nasiri MJ, Zamani S, et al. High genetic diversity among Mycobacterium tuberculosis strains in Tehran, Iran. J Clin Tuberc Other Mycobact Dis. 2018;11:1–6. doi:10.1016/j.jctube.2018.01.001
6. Molnar A. Antimicrobial resistance awareness and games. Trends Microbiol. 2018;27(1):1–3.
7. Tian L, Sun Z, Zhang Z. Antimicrobial resistance of pathogens causing nosocomial bloodstream infection in Hubei Province, China, from 2014 to 2016: a multicenter retrospective study. BMC Public Health. 2018;18(1):1121. doi:10.1186/s12889-018-6013-5
8. Shariati A, Azimi T, Ardebili A, et al. Insertional inactivation of oprD in carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Tehran, Iran. New Microbes New Infect. 2018;21:75–80. doi:10.1016/j.nmni.2017.10.013
9. Fahrenkamp-Uppenbrink J. Countering antibiotic resistance. Science. 2015;347(6226):1109–1111.
10. Maraki S, Mantadakis E, Mavromanolaki VE, Kofteridis DP, Samonis G. A 5-year surveillance study on antimicrobial resistance of Acinetobacter baumannii clinical isolates from a tertiary Greek hospital. J Infect Chemother. 2016;48(3):190–198. doi:10.3947/ic.2016.48.3.190
11. Aslam B. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–1658. doi:10.2147/IDR.S173867
12. Mahmoudi S, Mahzari M, Banar M, et al. Antimicrobial resistance patterns of Gram-negative bacteria isolated from bloodstream infections in an Iranian referral paediatric hospital: a 5.5-year study. J Glob Antimicrob Resist. 2017;11:17–22. doi:10.1016/j.jgar.2017.04.013
13. Zilberberg MD, Kollef MH, Shorr AF. Secular trends in Acinetobacter baumannii resistance in respiratory and blood stream specimens in the United States, 2003 to 2012: a survey study. J Int Med Res. 2016;11(1):21–26. doi:10.1002/jhm.2477
14. Nasser A, Jazireian P, Safari H, Alizade-Sani M, Pourmand MR, Azimi T. Staphylococcus aureus versus neutrophil: scrutiny of ancient combat. Microb Pathog. 2019;131:259–269. doi:10.1016/j.micpath.2019.04.026
15. Keihanian F, Saeidinia A, Abbasi K, Keihanian F. Epidemiology of antibiotic resistance of blood culture in educational hospitals in Rasht, North of Iran. Infect Drug Resist. 2018;11:1723. doi:10.2147/IDR.S169176
16. Bahramian A, Khoshnood S, Shariati A, Doustdar F, Chirani AS, Heidary M. Molecular characterization of the pilS2 gene and its association with the frequency of Pseudomonas aeruginosa plasmid pKLC102 and PAPI-1 pathogenicity island. Infect Drug Resist. 2019;12:221. doi:10.2147/IDR.S188527
17. Salehi B, Goudarzi H, Nikmanesh B, Houri H, Alavi-Moghaddam M, Ghalavand Z. Emergence and characterization of nosocomial multidrug-resistant and extensively drug-resistant Acinetobacter baumannii isolates in Tehran, Iran. J Int Med Res. 2018;24(7):515–523. doi:10.1016/j.jiac.2018.02.009
18. Maleki DT, Ghalavand Z, Laabei M, et al. Molecular analysis of accessory gene regulator functionality and virulence genes in Staphylococcus aureus derived from pediatric wound infections. Infect Genet Evol. 2019;73:255–260. doi:10.1016/j.meegid.2019.05.013
19. Tsutsui A, Suzuki S. Japan nosocomial infections surveillance (JANIS): a model of sustainable national antimicrobial resistance surveillance based on hospital diagnostic microbiology laboratories. BMC Health Serv Res. 2018;18(1):799. doi:10.1186/s12913-018-3604-x
20. Organization WH. Global action plan on antimicrobial resistance. Available from: https://www.who.int/antimicrobial-resistance/global-action-plan/en/
21. Organization WH. Global antimicrobial resistance surveillance system ( GLASS) report: early implementation 2017-2018. Available from: http://wwwwhoint/drugresistance/surveillance/GLASSmeeting/en/
22. Li L, Dai J-X, Xu L, et al. Antimicrobial resistance and pathogen distribution in hospitalized burn patients: a multicenter study in Southeast China. Medicine. 2018;97:34.
23. Jorak A, Keihanian F, Saeidinia A, Heidarzadeh A, Saeidinia F. A cross sectional study on knowledge, attitude and practice of medical students toward antibiotic resistance and its prescription, Iran. Adv Environ Biol. 2014;8(17):675–681.
24. Folgori L, Bielicki J, Heath PT, Sharland M. Antimicrobial-resistant Gram-negative infections in neonates: burden of disease and challenges in treatment. Curr Opin Infect Dis. 2017;30(3):281–288. doi:10.1097/QCO.0000000000000371
25. Jafari M, Fallah F, Borhan RS, et al. The first report of CMY, aac (6′)-Ib and 16S rRNA methylase genes among Pseudomonas aeruginosa isolates from Iran. Arch Pediatr Infec Dis. 2013;1(3):109–112. doi:10.5812/pedinfect.11392
26. Gopalakrishnan R, Sureshkumar D. Changing trends in antimicrobial susceptibility and hospital acquired infections over an 8 year period in a tertiary care hospital in relation to introduction of an infection control programme. J Assoc Physicians India. 2010;58(Suppl):25–31.
27. Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–318. doi:10.1179/2047773215Y.0000000030
28. Sedigh Ebrahim-Saraie H, Motamedifar M, Mansury D, Halaji M, Hashemizadeh Z, Ali-Mohammadi Y. Bacterial etiology and antibacterial susceptibility patterns of pediatric bloodstream infections: a two year study from Nemazee Hospital, Shiraz, Iran. J Compr Pediatr. 2016;7(1)):e29929. doi:10.17795/compreped-29929
29. Alam M, Pillai P, Kapur P, Pillai K. Resistant patterns of bacteria isolated from bloodstream infections at a university hospital in Delhi. J Pharm Bioallied Sci. 2011;3(4):525. doi:10.4103/0975-7406.90106
30. Buetti N, Atkinson A, Kottanattu L, Bielicki J, Marschall J, Kronenberg A. Patterns and trends of pediatric bloodstream infections: a 7-year surveillance study. Eur J Clin Microbiol Infect Dis. 2017;36(3):537–544. doi:10.1007/s10096-016-2830-6
31. Natsis NE, Cohen PR. Coagulase-negative Staphylococcus skin and soft tissue infections. Am J Clin Dermatol. 2018;19(5):671–677. doi:10.1007/s40257-018-0362-9
32. von Eiff C, Jansen B, Kohnen W, Becker K. Infections associated with medical devices. Drugs. 2005;65(2):179–214. doi:10.2165/00003495-200565020-00003
33. Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870–926. doi:10.1128/CMR.00109-13
34. Niederbracht Y, Idelevich E, Penner H, et al. Applicability of a commercial multiplex PCR test for identification of true blood stream infections with coagulase-negative staphylococci in neutropenic hematological patients.
35. Becker K, Skov RL, von Eiff C. Staphylococcus, micrococcus, and other catalase-positive cocci. In Jorgensen J, Pfaller M, Carroll K, Funke G, Landry M, Richter S, Warnock D, editors: Manual of Clinical Microbiology.
36. Hall KK, Lyman JA. Updated review of blood culture contamination. Clin Microbiol Rev. 2006;19(4):788–802. doi:10.1128/CMR.00062-05
37. Favre B, Hugonnet S, Correa L, Sax H, Rohner P, Pittet D. Nosocomial bacteremia clinical significance of a single blood culture positive for coagulase-negative staphylococci. Infect Control Hosp Epidemiol. 2005;26(8):697–702. doi:10.1086/502605
38. Dharmapalan D, Shet A, Yewale V, Sharland M. High reported rates of antimicrobial resistance in Indian neonatal and pediatric blood stream infections. J Pediatr Infect Dis Soc. 2017;6(3):e62–e68. doi:10.1093/jpids/piw092
39. He X, Xie M, Li S, et al. Antimicrobial resistance in bacterial pathogens among hospitalized children with community acquired lower respiratory tract infections in Dongguan, China (2011–2016). BMC Infect Dis. 2017;17(1):614. doi:10.1186/s12879-017-2757-2
40. Tian L, Sun Z, Zhang Z. Antimicrobial resistance of pathogens causing nosocomial bloodstream infection in Hubei Province, China, from 2014 to 2016: a multicenter retrospective study. BMC Public Health. 2018;18(1):1121.
41. Al-Naqshbandi AA, Chawsheen MA, Abdulqader HH. Prevalence and antimicrobial susceptibility of bacterial pathogens isolated from urine specimens received in rizgary hospital – Erbil. J Infect Public Health. 2018;12(3):330–336
42. Gandapor AJ, Khan AM. Antibiotic Sensitivity pattern of bacterial isolates of neonatal septicemia in Peshawar, Pakistan. Arch Iran Med. 2016;19(12):866.
43. Hui-min Y, Yan-ping W, Lin Liu Y, Shamsi BH, Bo H, Xu-chun M. Analysis of distribution and antibiotic resistance of pathogens isolated from the paediatric population in Shenmu Hospital from 2011–2015. J Int Med Res. 2018;46(1):225–233. doi:10.1177/0300060517716343
44. Moges F, Eshetie S, Yeshitela B, Abate E. Bacterial etiologic agents causing neonatal sepsis and associated risk factors in Gondar, Northwest Ethiopia. BMC Pediatr. 2017;17(1):137. doi:10.1186/s12887-017-0969-7
45. Behmadi H, Borji A, Taghavi-Rad A, Soghandi L, Behmadi R. Prevalence and antibiotic resistance of neonatal sepsis pathogens in Neyshabour, Iran. Arch Pediatr Infec Dis. 2016;4(2). doi:10.5812/pedinfect
46. Ardehali SH, Azimi T, Owrang M, Aghamohammadi N, Azimi L. Role of efflux pumps in reduced susceptibility to tigecycline in Acinetobacter baumannii. New Microbes New Infect. 2019;30:100547. doi:10.1016/j.nmni.2019.100547
47. Bahramian A, Shariati A, Azimi T, et al. First report of New Delhi metallo-beta-lactamase-6 (NDM-6) among Klebsiella pneumoniae ST147 strains isolated from dialysis patients in Iran. Infect Genet Evol. 2019;69:142–145. doi:10.1016/j.meegid.2019.01.030
48. Leclercq R, Canton R, Brown DF, et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect. 2013;19(2):141–160. doi:10.1111/j.1469-0691.2011.03703.x
49. Bassetti M, Righi E. New antibiotics and antimicrobial combination therapy for the treatment of gram-negative bacterial infections. Curr Opin Crit Care. 2015;21(5):402–411. doi:10.1097/MCC.0000000000000235
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
