Back to Journals » Infection and Drug Resistance » Volume 12
Molecular mechanisms related to colistin resistance in Enterobacteriaceae
Authors Aghapour Z, Gholizadeh P , Ganbarov K , Bialvaei AZ , Mahmood SS, Tanomand A , Yousefi M , Asgharzadeh M, Yousefi B , Kafil HS
Received 29 December 2018
Accepted for publication 4 March 2019
Published 24 April 2019 Volume 2019:12 Pages 965—975
DOI https://doi.org/10.2147/IDR.S199844
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Zahra Aghapour,1,2 Pourya Gholizadeh,3 Khudaverdi Ganbarov,4 Abed Zahedi Bialvaei,5 Suhad Saad Mahmood,6 Asghar Tanomand,7 Mehdi Yousefi,8 Mohammad Asgharzadeh,9 Bahman Yousefi,9 Hossein Samadi Kafil1
1Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran; 2Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran; 3Tuberculosis and Lung Diseases Research Center, Tabriz University of Medical Sciences, Tabriz, Iran; 4Department of Microbiology, Baku State University, Baku, Azerbaijan; 5Department of Microbiology, Iran University of Medical Sciences, Tehran, Iran; 6Department of Biotechnology, College of Science, University of Baghdad, Baghdad, Iraq; 7Department of Microbiology, Maragheh University of Medical Sciences, Maragheh, Iran; 8Stem Cell Research Center, Tabriz University of Medical Sciences, Tabriz, Iran; 9Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract: Colistin is an effective antibiotic for treatment of most multidrug-resistant Gram-negative bacteria. It is used currently as a last-line drug for infections due to severe Gram-negative bacteria followed by an increase in resistance among Gram-negative bacteria. Colistin resistance is considered a serious problem, due to a lack of alternative antibiotics. Some bacteria, including Pseudomonas aeruginosa, Acinetobacter baumannii, Enterobacteriaceae members, such as Escherichia coli, Salmonella spp., and Klebsiella spp. have an acquired resistance against colistin. However, other bacteria, including Serratia spp., Proteus spp. and Burkholderia spp. are naturally resistant to this antibiotic. In addition, clinicians should be alert to the possibility of colistin resistance among multidrug-resistant bacteria and development through mutation or adaptation mechanisms. Rapidly emerging bacterial resistance has made it harder for us to rely completely on the discovery of new antibiotics; therefore, we need to have logical approaches to use old antibiotics, such as colistin. This review presents current knowledge about the different mechanisms of colistin resistance.
Keywords: colistin, Enterobacteriaceae, two-component system, lipid A, mcr genes
Introduction
Antibiotic resistance, which started in the 1970s among Gram-negative bacteria, is a crucial global problem.1–3 Development of antibiotic resistance is a phenomenon correlated with antibiotic overuse and bacterial evolution.4 Microorganisms can use several mechanisms to adapt against antimicrobial agents and environmental stimulants. Bacteria can use genetic alterations in their genes to form genes with improved performance to overcome antibiotics. Modification in only a few base pairs in DNA causing replacement of one or a few amino acids in an important target, such as cell structure or cell wall and enzymes, leads to new resistance strains.5 Initially, the problem of bacterial resistance to antibiotics was solved by the invention of the latest categories of antibiotics, including aminoglycosides, glicopeptides, and macrolides, and further by the c
hemical modification of old antibiotics. Unfortunately, these antibiotics could not keep pace with the development of antibiotic resistance in bacterial pathogens.6 Mobile genes conferring resistance to aminoglycosides and broad-spectrum β-lactams can transfer between species and are one of the important factors accounting for the progressive erosion of antimicrobial activity in both hospital and community settings.7 Emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) Gram-negative bacteria, as well as the lack of novel agents against these pathogens, have led to the reintroduction of colistin, an old and valuable antibiotic as a last-resort treatment option.8
Colistin, also known as polymyxin E, was isolated in 1947 from the bacterium Paenibacillus polymyxa subsp. colistinus.9 This organism also produces colistinase, which inactivates colistin.10 Colistin is a polycationic antibiotic, and has significant activity against Gram negative bacteria, such as Enterobacteriaceae. The outer cell membrane of Gram-negative bacteria is the main site of action for colistin. When colistin binds to lipopolysaccharides in the outer membrane, electrostatic interaction occurr between the α,γ-diaminobutyric acid of colistin and the phosphate groups of the lipid A region of lipopolysaccharide (LPS). It competitively displaces divalent cations (Ca2+ and Mg2+) from the phosphate groups of membrane lipids.11,12 Therefore, disruption of LPS may cause increased permeability of the outer membrane and leakage of intracellular contents, ultimately leading to cell death.13–15 Unfortunately, during the last few decades, the emergence of colistin-resistant isolates has been frequently reported,10,12 which has increased inappropriate use of this drug, especially as monotherapy could be the cause of this problem.16–18 In addition, there have been reports of increased infection due to bacteria with intrinsic resistance to colistin, such as Proteus spp., Providencia spp., Serratia spp., and Morganella spp.19–21 In this article, we assess different mechanisms of colistin resistance in Enterobacteriaceae.
Activity spectrum of colistin
Colistin is a narrow-spectrum antimicrobial agent that has significant activity against most members of the Enterobacteriaceae family, including Escherichia coli, Enterobacter spp., Klebsiella spp., Citrobacter spp., Salmonella spp., and Shigella spp. It also has activity against common nonfermentative Gram-negative bacteria, such as Acinetobacter baumannii, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia.14,22–24 In addition, Haemophilus influenzae, Legionella pneumophila, Aeromonas spp., and Bordetella pertussis are naturally susceptible to colistin.15,22,25,26
Conversely, among the Enterobacteriaceae, Proteus spp. and Serratia marcescens have intrinsic resistance to colistin. On the other hand, Morganella morganii, Providencia spp., Pseudomonas mallei, Burkholderia cepacia, Chromobacterium spp., Edwardsiella spp., Brucella, Legionella, and Vibrio cholera are typically resistant to colistin. Colistin is not active against Gram-negative cocci, such as Neisseria spp., gramGram-positive bacteria, anaerobic bacteria, eukaryotic microbes, or mammalian cells.14,27–31
Mechanisms of colistin resistance in Enterobacteriaceae
Although the main mechanism of resistance to colistin is unclear, Gram-negative bacteria employ several mechanisms to protect themselves against colistin toward other polymyxins (Figure 1). According to the literature, most colistin-resistance mechanisms are adaptive mechanisms ithat occur after in vitro exposure.15 Resistance to colistin occur with LPS modification via different routes. The most common strategies for resistance to colistin are modifications of the bacterial outer membrane through alteration of the LPS and reduction in its negative charge.32,33 The other strategy is the overexpression of efflux-pump systems.34 Another mechanism is overproduction of capsule polysaccharide.35–37 No enzymatic mechanisms of resistance have been reported, but strains of P. polymyxa produce colistinase.38
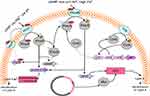 | Figure 1 Regulation and plasmid-mediated pathways of lipopolysaccharide modifications in Enterobacteriaceae. |
Intrinsic resistance mechanisms
Resistance to polymyxins occurs naturally in P. mirabilis and S. marcesens by modification of the LPS via cationic substitution. The mechanism of resistance in these species is linked to expression of the arnBCADTEF operon and the eptB gene. In this way, the 4-amino-4-deoxy-L-arabinose (L-Ara4N) and phosphoethanolamine (pEtN) cationic groups are added to the LPS by this operon and gene, respectively. It has been shown that the LPS of P. mirabilis contains L-Ara4N and the genome of this bacterium contains the eptC gene, which is mediated to the modification of LPS with PETN.39–41 Putative loci in P. mirabilis include the sap operon encoding a transport protein, ATPase gene, and O-acetyltransferase gene, which take part in biosynthesis or transfer of amino arabinose.42 Also, the existence of rppA/rppB TCS has been discovered to play a role in activation of the arnBCADTEF operon.43,44 Similarly, this operon is responsible for intrinsic resistance to colistin in S. marcescens, as it has been shown that arnB and arnC mutants lead to a reduction insusceptibility to colistin (minimum inhibitory concentration [MIC] from 2,048 to 2 µg/mL) compared to the wild type.45
This modification of LPS and the increase inits charge give rise to the affinity of colistin decrease for binding to LPS. Therefore, intrinsic resistance has occurred in these species.9,41,43
Acquired resistance mechanisms in Enterobacteriaceae
Acquired colistin-resistance mechanisms have been recognized in some members of Enterobacteriaceae family, such as E. coli, Salmonella spp., Klebsiella spp., and Enterobacter spp., and remain unknown for other bacterial species. Resistance mechanisms are presumed to be linked to chromosomal mutation untransferable via horizontal gene transfer.46–48 Only one mechanism of resistance has been identified as a transferable mechanism (plasmid-mediated mcr gene) so far (Table 1).9,21
 | Table 1 Acquired and intrinsic strategies employed by Gram-negative bacteria for achieving resistance to colistin |
Many genes and operons play a role in modification of LPS, which in turn leads to colistin resistance. These include: genes and operons responsible for encoding enzymes that have a direct role in LPS modification, such as the pmrC and pmrE genes and the pmrHFIJKLM operon;46,49 regulatory two-component systems (TCSs), including PmrAB and PhoPQ, as well as crrAB, which regulates the PmrAB system;50–52 the mgrB gene, a negative regulator of TCSs, including PmrAB and PhoPQ;53 plasmid-mediated mcr genes;54,55 and Cpx and Rcs as regulator of upregulation of capsule biosynthesis and activator of the efflux pump KpnEF regulating the PhoPQ system, respectively.8
mgrB gene and regulators of PmrAB and PhoPQ two-component systems
Some operons and regulators have a role in the modification of LPS by PmrAB and PhoPQ TCSs. The pmrABC operon encodes PmrA (BasR) as a regulator protein, PmrB (BasS) as a cytoplasmic membrane-bound sensor kinase, and PmrC as a putative membrane protein.56 The addition of L-arabinoseamine (L-Ara4N) to the 1-phosphate or 4́ -phosphate group leads to colistin resistance.46 Generally, L-Ara4N is connected to 4́-phosphate and modifies it while PETN is connected to 1-phosphate.57,58 The pmrHFIJKLM operon (also named arnBCDADTEF or pbgPE) and PmrE synthesize L-Ara4N from uridine diphosphate glucuronic acid and fix it to lipid A.59,60 The biosynthesis of L-Ara4N depends on the pmr (arn) operon.61 Moreover, under environmental stimulants, such as macrophage phagosomes, the high concentration of iron (Fe3+) and exposure to aluminum (Al3+), as well as acidic pH, leads to activation of PmrB.56,62 On the other hand, low concentration of Mg2+ or Ca2+ leads to activation of phoQ.63,64 PmrB activates PmrA by phosphorylation, and PmrA in turns activates regulation of the pmrABC and pmrHFIJKLM operons and the pmrE gene. Subsequently, these operons and genes lead to LPS modification by adding PETN and L-Ara4N to lipid A.56 Mutation of pmrA/pmrB results in upregulation of the pmrABC and pmrFHIJKLM operons and pmrE gene. Mutation within the pmrA and pmrB genes leading to colistin resistance has been described in Klebsiella pneumoniae and Salmonella entericca (Table 1).65–69
On the other hand, the phoPQ TCS encodes PhoP as a regulator protein and PhoQ as a sensor kinase. Under conditions of low magnesium or calcium, acidic PH, or cationic antimicrobial peptide, PhoPQ is activated and protects bacteria.21,63,64 Activated PhoPQ leads to modification of lipid A via two routes: PhoQ activates PhoP by its kinase activity via phosphorylation, which activates transcription of the pmrFHIJKLM operon, followed by modification of lipid A;70,71 and PhoP indirectly activates pmrA by bypassing the PmrD connector protein, subsequently activates the transcription of the pmrHFIJKLM operon and synthesizes PETN, which transfers it to lipid A.72,73
Various of PETN-coding genes, such as eptA (pmrC), eptB (pagC), and eptC (cptA), are able to add PETN to different sites of LPS.74,75 Mutation of the phoP/Q genes has been identified in K. pneumoniae and E. coli that led to acquired colistin resistance.65,67,76–78
The mgrB gene encodes a small transmembrane protein of 47 amino acids that exerts negative feedback on the PhoPQ TCS.79 This protein inhibits the kinase activity of PhoQ, which in turn represses expression of the phoQ gene. Nevertheless, mutation/inactivation of the mgrB gene results in upregulation of the phoPQ operon and subsequent activation of the pmrHFIJKLM operon. Finally, production of L-Ara4N leads to modification of lipid A and colistin resistance.51
Various mutations or disruptions of the mgrB gene have been reported, such as deletion, nonsense, missense, inactivation, and insertional mutations. According to reports, mgrB inactivation is the most common mechanism for colistin resistance in K. pneumoniae and K. oxytoca.67,80–82 In addition, it has been described that inactivation of the mgrB gene by diverse insertion sequences at different sites of this gene is the other mgrB mutation that often occurs in K. pneumoniae.53,65,80 Other alterations that have been reported in the mgrB gene include nonsense and missense mutations, leading to premature termination and amino-acid substitutions inmgrB, respectively.53,77 Goulian et al showed that deletion of the mgrB gene led to upregulation of the PhoP-regulated gene in E. coli.79
CrrAB two-component system
The crrAB operon encodes two proteins: CrrA as a regulatory protein and CrrB as a sensor kinase protein. Wright et al described that mutation of crrB leads to colistin resistance in K. pneumoniae.83 The mutated CrrB protein regulates a crrAB-adjacent gene that encodes a glycosyltransferase-like protein, which in turn leads to modification of lipid A.83 In Cheng et al's study, six amino-acid substitutions in the CrrB protein led to high resistance to colistin (MICs of colistin 512–2,048 µg/mL).52 However, mutation/inactivation of the crrB gene led to activation of the pmrHFIJKLM operon and the pmrC and pmrE genes through overexpression of the pmrAB operon. Furthermore, the production and addition of L-Ara4N and PETN to lipid A lead to acquisition of resistance to colistin.83 It was demonstrated that CrrC afforded a connection between the CrrAB and pmrAB systems. Mutation of the crrB gene led to increased crrC transcription. On the other hand, it has been suggested amino-acid substitutions of the CrrB protein result in increased autophosphorylation of this protein, consequently leading to colistin resistance.52
Plasmid-mediated resistance to colistin
Plasmid-mediated colistin is a significant challenge and global concern, because of easy transfer of colistin-resistance genes to susceptible strains.54 The mcr genes are responsible for horizontal transfer of colistin resistance. These plasmid-mediated genes were first reported in E. coli isolated from pigs and meat in China, November 2015.54 MCR is a member of the PETN enzyme family, and its expression leads to addition of PETN to lipid A. According to the literature, isolates carrying the mcr1 gene display resistance to colistin without other resistance mechanisms. The existence of mcr1 in isolates is enough for colistin resistance without other resistance mechanisms, as isolates carrying this gene displayed a four- to eightfold increase in colistin MIC.9 It is worth noting that the production of mcr1 leads to resistance to lysozymes.84
Following initial findings, mcr1-mediating transferable colistin resistance has been reported in several regions, including Europe, Asia, the Americas, and Africa.85–98 There is a hypothesis that mcr1 originated in animals, particularly pigs and cattle, and subsequently spread to humans, though the proportion of mcr1-positive isolates is low in humans compared to animals.54,99 This transmissible gene has been reported from diverse genera of Enterobacteriaceae, including E. coli, Klebsiella spp., Entrobacter spp., Salmonella spp., Shigella spp., and Cronobacter spp., but mostly from E. coli. Some plasmids containing the mcr1 gene carry other genes that are resistant to other antibiotics, such as β-lactams, aminoglycosides, quinolones, sulfonamides, tetracyclines, and fosfomycin.9 The mcr gene has also been identified in Enterobacteriaceae isolates, which carry such carbapenemase genes as bla
Recently, Xavier et al reported a novel plasmid-mediated colistin resistance gene, known as mcr2, in E. coli.55 Thereafter, mcr3 and mcr4 genes were discovered.102,103 Finally, in July 2017, Borowiak et al reported a new gene of the mcr family from Salmonella paratyphi B were carried in transposons instead of plasmids.104
In addition, three mobile colistin-resistance genes (mcr6, mcr7, and mcr8) were discovered in 2018. AbuOun et al discovered a new variant of mcr2 from Moraxella pluranimalium that they renamed mcr6.1.105 They suggested that Moraxella spp. may contain a natural reservoir of mcr, and mcr-harboring Moraxella appeared in pig populations. Yang et al found K. pneumoniae isolates harbored a new mcr variant, mcr7.1, recovered from chickens in China.106 They suggested that mcr7, like mcr-3, originated from Aeromonasspp.,102 and its structure was similar to mcr3. In addition, mcr7 displayed 78% nucleotide identity to the mcr3 gene. Eventually, a new mobile genetic element, mcr-8, was discovered in K. pneumoniae. It was identified as the coexistence of mcr8 and the carbapenemase-encoding gene blaNDM, which is a great concern.107 It is notable that mcr8 has existed for some time and disseminated among K. pneumoniae.107 mcr2–8 are similar to mcr1, as PETN leads to the addition of phosphoethanolamine to lipid A, followed by colistin resistance (Figure 1). Both mcr1 and mcr2 genes originated from Moraxella spp. In addition, mcr3 and mcr4 genes line up closely with PETN from Aeromonas spp. and Shewanella frigidimarina, respectively,55,102,103,108 whereas the origin of mcr5 remains unknown.104 Although mcr is a plasmid-mediated gene, recently Zurfluh et al identified the mcr1 gene on chromosomes of E. coli strains. Therefore, there is a hypothesis that this gene can be integrated in the genome of some isolates.109
Role of regulator RamA
The ramA locus has three genes: ramA, romA, and ramR. The ramR gene plays a role as a repressor of the ramA and romA genes. Some Enterobacteriaceae possess a ramA regulator, such as K. pneumoniae, Citrobacter spp., Enterobacter spp., and Salmonella spp. In K. pneumoniae, this regulator modulates lipid A biosynthesis and is related to permeability barriers. It has been shown that ramA alterations lead to reductions in colistin susceptibility. Recently, researchers showed that increased levels of RamA resulted in LPS modification and increased resistance to colistin.110 RamA applied changes tothe bacterial surface and Klebsiella survived against colistin. Several genes are associated with lipid A biosynthesis, including lpxA, lpxC, lpxD, lpxB, lpxK, lpxL, lpxM, and lpxO.111 RamA binds directly to and activates the lpxC, lpxO, and lpxL2 genes and leads to alterations within the lipid A moiety in K. pneumoniae. Therefore, Klebsiella can survive in such antibiotic challenges as colistin.110
Role of capsule in colistin resistance
The role of capsular polysaccharide (CPS) has been demonstrated to be protective against cationic antimicrobial peptides, including colistin.35 K. pneumoniae is able to release CPS from its surface.112 The number of capsule layers is related to resistance level. It has been observed that K. pneumoniae with several layers was more resistant to colistin than isolates with few layers.8,113 However, upregulation of a capsular biosynthesis gene led to a reduction in the interaction of colistin with the target site in K. pneumoniae, followed by increased colistin resistance.35 Consequently, there are some regulators of capsule formation, such as Cpx (conjugative pilus expression) and Rcs (regulator of capsule synthesis). Cpx and Rcs also appear to contribute to colistin resistance by activating the efflux pump KpnEF and regulating the PhoPQ TCS, respectively.46 Furthermore, the ugd gene plays a role in CPS and L-Ara4N biosynthesis in that its phosphorylation is related to the synthesis of capsular and colistin resistance.114,115
Role of efflux pumps
A few studies have suggested that efflux-pump systems are involved in colistin resistance. Efflux pumps, such as the KpnEF, AcrAB and Sap proteins, have been reported in Enterobactericeae. By activation of these pumps, resistance to colistin is increased.116,117 The efflux pump KpnEF is a member of the Cpx regulon (responsible for capsule synthesis in K. pneumoniae) and belongs to the SMR protein family.8 In K. pneumoniae, this pump is mediated by colistin resistance and other antibiotics, including ceftriaxone, erythromycin, and rifampicin.117 It has been observed that mutations in KpnEF (as a member of the small MDR efflux-pump family) lead to more susceptibility and a doubled reduction inthe MIC of colistin.117 On the other hand, AcrAB is a part of the AcrAB–TolC complex, which plays a role in colistin resistance. The AcrAB-mutant E. coli displays aneightfold increase in colistin susceptibility. It has been remarked that expression of this pump's proteins is dependent on the PhoPQ TCS.118 Finally, the SapABCDF operon encodes Sap proteins that are constitute of five proteins.118 In the mutant of P. mirabilis, susceptibility to colistin is increased by mutation of the SapABCDEF operon.42 It has been shown that the use of efflux-pump inhibitors in the test medium carbonyl cyanide 3-chlorophenylhydrazone leads to a reduction in MIC for colistin-resistant strains.119
Logical approaches to use of colistin
Recent studies have suggested colistin is the foremost therapeutic option of XDR Gram-negative bacteria in recent years, owing to its potent bactericidal efficacy.120 Combination therapies of colistin with other antibiotics are superior to colistin monotherapy for XDR strains, due to rapid selection of resistance in some strains, heteroresistance during colistin monotherapy, and lower clinical efficacy during colistin-based combination.121 In addition, rates of cure, 14-day survival, and microbiological eradication are lower in monotherapy compared to combination therapy.121 Moreover, several combination therapies have been recommended to decrease the development of resistance. The combination of colistin with other drugs, such as carbapenems, sulbactam, tigecycline, aminoglycosides, and rifampicin, has been recommended to prevent the development of colistin-resistant strains, which may improve clinical and microbiological outcomes.121–126 The colistin–sulbactam combination was recommended against imipenem-resistant A. baumannii, particularly in colistin-resistant strains, due to its high in vitro synergistic activity,121,127 which may be a more favorable combination. Colistin-based combinations with tigecycline, aminoglycosides, and rifampicin have shown synergistic activity against XDR strains,122,125,128 but tigesycline is disadvantageous in bacteremic patients, because of its low plasma concentrations.128 In addition, colistin–carbapenem combinations may be preferable in the treatment of A. baumannii infections to prevent resistance selection and to decrease the prevalence of A. baumannii.121
Conclusion
The main target for colistin is lipid A of the LPS in Gram-negative bacteria, leading to disruption of the bacterial membrane and resulting in cellular death. In recent decades, the increasing use of colistin in clinical settings, mainly in veterinary clinics, has led to the emergence of colistin resistance. Many studies have shown that the prevalence of colistin resistance has increased rapidly among Enterobacteriaceae. Clinicians should be alert to the possibility of colistin resistance among MDR bacteria and the development of colistin resistance through mutation or adaptation mechanisms. Rapidly emerging bacterial resistance has made it harder for us to rely completelyon the discovery of new antibiotics; therefore, we need to have logical approaches to use older antibiotics, such as colistin.
Acknowledgments
This study received no funding, and was the authors' own work. We thank the staff of the Drug Applied Research Center for their support.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin Infect Dis. 2009;48(1):1–12. doi:10.1086/595011
2.
3. Shlaes DM, Sahm D, Opiela C, Spellberg B. chemotherapy. Commentary: the FDA reboot of antibiotic development. Antimicrob Agents Chemother. 2013;57(10):4605-4607.
4. Talbot GH, Bradley J, Edwards JE
5. Tenover FC, McGowan JE
6. Gold HS, Moellering RC
7. Jeannot K, Bolard A, Plésiat PJI. Resistance to polymyxins in gram-negative organisms. Int J Antimicrob Agents. 2017;49(5):526–535. doi:10.1016/j.ijantimicag.2016.11.029
8. Baron S, Hadjadj L, Rolain J-M, Olaitan AO. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents. 2016;48(6):583–591. doi:10.1016/j.ijantimicag.2016.06.023
9. Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30(2):557–596. doi:10.1128/CMR.00064-16
10. Yahav D, Farbman L, Leibovici L, Paul M. Colistin: new lessons on an old antibiotic. Clin Microbiol Infect. 2012;18(1):18–29. doi:10.1111/j.1469-0691.2011.03734.x
11. Dixon RA, Chopra I. Leakage of periplasmic proteins from Escherichia coli mediated by polymyxin B nonapeptide. Antimicrob Agents Chemother. 1986;29(5):781–788.
12. Bialvaei AZ, Samadi Kafil H. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin. 2015;31(4):707–721. doi:10.1185/03007995.2015.1018989
13. Li J, Nation RL, Turnidge JD, et al. Colistin: the re-emerging antibiotic for multidrug-resistant gram-negative bacterial infections. Lancet Infect Dis. 2006;6(9):589–601. doi:10.1016/S1473-3099(06)70580-1
14. Falagas ME, Kasiakou SK, Saravolatz LD. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis. 2005;40(9):1333–1341. doi:10.1086/429323
15. Biswas S, Brunel J-M, Dubus J-C, Reynaud-Gaubert M, Rolain J-M. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther. 2012;10(8):917–934. doi:10.1586/eri.12.78
16. Capone A, Giannella M, Fortini D, et al. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect. 2013;19(1):E23–E30. doi:10.1111/1469-0691.12070
17. Lee J-Y, Ko KS. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn Microbiol Infect Dis. 2014;78(3):271–276. doi:10.1016/j.diagmicrobio.2013.11.027
18. Bialvaei AZ, Kafil HS, Asgharzadeh M, Yousef Memar M, Yousefi M. Current methods for the identification of carbapenemases. J Chemother. 2016;28(1):1–19. doi:10.1179/1973947815Y.0000000063
19. Hayakawa K, Marchaim D, Divine GW, et al. Growing prevalence of Providencia stuartii associated with the increased usage of colistin at a tertiary health care center. Int J Infect Dis. 2012;16(9):e646–e648. doi:10.1016/j.ijid.2012.05.1029
20. Samonis G, Korbila I, Maraki S, et al. Trends of isolation of intrinsically resistant to colistin Enterobacteriaceae and association with colistin use in a tertiary hospital. Eur J Clin Microbiol Infect Dis. 2014;33(9):1505–1510. doi:10.1007/s10096-014-2097-8
21. Aghapour Z, Hasani A, Aghazadeh M, et al. Genes involved in colistin resistance of gram-negative isolates in the northwest of Iran. Gene Rep. 2019;14:81–86. doi:10.1016/j.genrep.2018.12.001
22. Li J, Nation RL, Milne RW, Turnidge JD, Coulthard K. Evaluation of colistin as an agent against multi-resistant gram-negative bacteria. Int J Antimicrob Agents. 2005;25(1):11–25. doi:10.1016/j.ijantimicag.2004.10.001
23. Tan T, Ng S. The in-vitro activity of colistin in gram-negative bacteria. Singapore Med J. 2006;47(7):621.
24. Bialvaei AZ, Kouhsari E, Salehi-Abargouei A, et al. Epidemiology of multidrug-resistant Acinetobacter baumannii strains in Iran: a systematic review and meta-analysis. J Chemother. 2017;29(6):327–337. doi:10.1080/1120009X.2017.1338377
25. Giamarellou H, Poulakou G. Multidrug-resistant gram-negative infections. Drugs. 2009;69(14):1879–1901. doi:10.2165/11315690-000000000-00000
26. Gales AC, Jones R, Sader HS. Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of gram-negative bacilli: report from the SENTRY antimicrobial surveillance programme (2001–2004). Clin Microbiol Infect. 2006;12(4):315–321. doi:10.1111/j.1469-0691.2005.01351.x
27. Gales AC, Jones RN, Sader HS. Contemporary activity of colistin and polymyxin B against a worldwide collection of gram-negative pathogens: results from the SENTRY antimicrobial surveillance program (2006–09). J Antimicrob Chemother. 2011;66(9):2070–2074. doi:10.1093/jac/dkr239
28. Vaara M. Polymyxins and their novel derivatives. Curr Opin Microbiol. 2010;13(5):574–581. doi:10.1016/j.mib.2010.09.002
29. Muyembe T, Vandepitte J, Desmyter J. Natural colistin resistance in Edwardsiella tarda. Antimicrob Agents Chemother. 1973;4(5):521–524.
30. Shimizu S, Iyobe S, Mitsuhashi S. Inducible high resistance to colistin in Proteus strains. Antimicrob Agents Chemother. 1977;12(1):1–3.
31. Bialvaei AZ, Kafil HS, Asgharzadeh M. Role of treatment cost on transmission of multidrug-resistant tuberculosis into Iran. Clin Infect Dis. 2015;61(6):1029–1030. doi:10.1093/cid/civ459
32. Landman D, Georgescu C, Martin DA, Quale J. Polymyxins revisited. Clin Microbiol Rev. 2008;21(3):449–465. doi:10.1128/CMR.00006-08
33. Nation RL, Li J. Colistin in the 21st century. Curr Opin Infect Dis. 2009;22(6):535. doi:10.1097/QCO.0b013e328332e672
34. Bengoechea JA, Skurnik M. Temperature‐regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol Microbiol. 2000;37(1):67–80.
35. Campos MA, Vargas MA, Regueiro V, Llompart CM, Albertí S, Bengoechea JA. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun. 2004;72(12):7107–7114. doi:10.1128/IAI.72.12.7107-7114.2004
36. Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2010;54(1):177–183. doi:10.1128/AAC.00715-09
37. Kim Y, Bae IK, Lee H, Jeong SH, Yong D, Lee K. In vivo emergence of colistin resistance in Acinetobacter baumannii clinical isolates of sequence type 357 during colistin treatment. Diagn Microbiol Infect Dis. 2014;79(3):362–366. doi:10.1016/j.diagmicrobio.2014.03.027
38. Ito-Kagawa M, Koyama Y. Selective cleavage of a peptide antibiotic, colistin by colistinase. J Antibiot. 1980;33(12):1551–1555.
39. Sidorczyk Z, Zähringer U, Rietschel ET. Chemical structure of the lipid A component of the lipopolysaccharide from a Proteus mirabilis Re-mutant. Eur J Biochem. 1983;137(1‐2):15–22.
40. Boll M, Radziejewska-Lebrecht J, Warth C, Krajewska-Pietrasik D, Mayer H. 4-Amino-4-deoxy-L-arabinose in LPS of enterobacterial R-mutants and its possible role for their polymyxin reactivity. FEMS Immunol Med Microbiol. 1994;8(4):329–341.
41. Aquilini E, Merino S, Knirel YA, Regué M, Tomás JM. Functional identification of Proteus mirabilis eptC gene encoding a core lipopolysaccharide phosphoethanolamine transferase. Int J Mol Sci. 2014;15(4):6689–6702. doi:10.3390/ijms15046689
42. McCoy AJ, Liu H, Falla TJ, Gunn JS. Identification of Proteus mirabilisMutants with increased sensitivity to antimicrobial peptides. Antimicrob Agents Chemother. 2001;45(7):2030–2037. doi:10.1128/AAC.45.7.2030-2037.2001
43. Jiang -S-S, Liu M-C, Teng L-J, Wang W-B, Hsueh P-R, Liaw S-J. Proteus mirabilis pmrI, an RppA-regulated gene necessary for polymyxin B resistance, biofilm formation, and urothelial cell invasion. Antimicrob Agents Chemother. 2010;54(4):1564–1571. doi:10.1128/AAC.01219-09
44. Wang W-B, Chen I-C, Jiang -S-S, et al. Role of RppA in the regulation of polymyxin b susceptibility, swarming, and virulence factor expression in Proteus mirabilis. Infect Immun. 2008;76(5):2051–2062. doi:10.1128/IAI.01557-07
45. Lin QY, Tsai Y-L, Liu M-C, Lin W-C, Hsueh P-R, Liaw S-J. Serratia marcescens arn, a PhoP-regulated locus necessary for polymyxin B resistance. Antimicrob Agents Chemother. 2014;58(9):5181–5190.
46. Olaitan AO, Morand S, Rolain J-M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi:10.3389/fmicb.2014.00547
47. Lee J-Y, Choi M-J, Choi HJ, Ko KS. Preservation of acquired colistin resistance in gram-negative bacteria. Antimicrob Agents Chemother. 2016;60(1):609–612. doi:10.1128/AAC.01574-15
48. Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1):42. doi:10.1038/nrmicro3380
49. Falagas ME, Rafailidis PI, Matthaiou DK. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Updat. 2010;13(4–5):132–138. doi:10.1016/j.drup.2010.05.002
50. Jayol A, Nordmann P, Brink A, Poirel L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother. 2015;59(5):2780–2784.
51. Cannatelli A, D’Andrea MM, Giani T, et al. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemase mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother. 2013;57(11)5521–5526.
52. Cheng Y-H, Lin T-L, Lin Y-T, Wang J-T. Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2016;60(6):3709–3716.
53. Cannatelli A, Giani T, D’Andrea MM, et al. MgrB inactivation is a common mechanism of colistin resistance in KPC carbapenemase-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother. 2014;58(10):5696–5703.
54. Liu -Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
55. Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27):30280. doi:10.2807/1560-7917.ES.2016.21.27.30280
56. Gunn JS. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 2008;16(6):284–290. doi:10.1016/j.tim.2008.03.007
57. Zhou Z, Ribeiro AA, Lin S, Cotter RJ, Miller SI, Raetz CR. Lipid a modifications in polymyxin resistant Salmonella typhimurium: PmrA dependent 4-amino-4-deoxy-L-arabinose and phosphoethanolamine incorporation. J Biol Chem. 2001;276(46):43111–43121.
58. Helander IM, Kilpeläinen I, Vaara M. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of the polymyxin-resistant pmrA mutants of Salmonella typhimurium: a 31P-NMR study. Mol Microbiol. 1994;11(3):481–487.
59. Gatzeva-Topalova PZ, May AP, Sousa MC. Structure and mechanism of ArnA: conformational change implies ordered dehydrogenase mechanism in key enzyme for polymyxin resistance. Structure. 2005;13(6):929–942. doi:10.1016/j.str.2005.03.018
60. Yan A, Guan Z, Raetz CR. An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J Biol Chem. 2007;282:36077–36089. doi:10.1074/jbc.M706172200
61. Reeves PR, Hobbs M, Valvano MA, et al. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4(12):495–503.
62. McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(1):205–217.
63. Gunn JS, Miller SI. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178(23):6857–6864.
64. Moskowitz SM, Ernst RK, Miller SI. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol. 2004;186(2):575–579.
65. Nordmann P, Jayol A, Poirel L. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis. 2016;22(6):1038. doi:10.3201/eid2206.151840
66. Cannatelli A, Di Pilato V, Giani T, et al. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC carbapenemase-producing Klebsiella pneumoniae associated with low-dosage colistin treatment. Antimicrob Agents Chemother. 2014;58(8):4399–4403.
67. Cheng Y-H, Lin T-L, Pan Y-J, Wang Y-P, Lin Y-T, Wang J-T. Colistin-resistant mechanisms of Klebsiella pneumoniae in Taiwan. Antimicrob Agents Chemother. 2015;59(5):2909–2913.
68. Diene SM, Merhej V, Henry M, et al. The rhizome of the multidrug-resistant Enterobacter aerogenes genome reveals how new “killer bugs” are created because of a sympatric lifestyle. Mol Biol Evol. 2012;30(2):369–383. doi:10.1093/molbev/mss236
69. Olaitan AO, Dia NM, Gautret P, et al. Acquisition of extended-spectrum cephalosporin-and colistin-resistant Salmonella enterica subsp. enterica serotype newport by pilgrims during Hajj. Int J Antimicrob Agents. 2015;45(6):600–604. doi:10.1016/j.ijantimicag.2015.01.010
70. Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183(6):1835–1842. doi:10.1128/JB.183.6.1835-1842.2001
71. Park SY, Groisman EA. Signal-specific temporal response by the S almonella PhoP/PhoQ regulatory system. Mol Microbiol. 2014;91(1):135–144. doi:10.1111/mmi.12449
72. Fu W, Yang F, Kang X, et al. First structure of the polymyxin resistance proteins. Biochem Biophys Res Commun. 2007;361(4):1033–1037. doi:10.1016/j.bbrc.2007.07.144
73. Cheng H-Y, Chen Y-F, Peng H-L. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci. 2010;17(1):60. doi:10.1186/1423-0127-17-74
74. Park YK, Lee J-Y, Ko KS. Transcriptomic analysis of colistin-susceptible and colistin-resistant isolates identifies genes associated with colistin resistance in Acinetobacter baumannii. Clin Microbiol Infect. 2015;21(8):
75. Qureshi ZA, Hittle LE, O’hara JA, et al. Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin Infect Dis. 2015;60(9):1295–1303. doi:10.1093/cid/civ048
76. Choi M-J, Ko KS. Mutant prevention concentrations of colistin for Acinetobacter baumannii, Pseudomonas aeruginosa and Klebsiella pneumoniae clinical isolates. J Antimicrob Chemother. 2013;69(1):275–277. doi:10.1093/jac/dkt315
77. Olaitan AO, Diene SM, Kempf M, et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents. 2014;44(6):500–507. doi:10.1016/j.ijantimicag.2014.07.020
78. Olaitan AO, Thongmalayvong B, Akkhavong K, et al. Clonal transmission of a colistin. Microb Drug Resist. 2014;20:310–315. doi:10.1089/mdr.2013.0193
79. Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009;5(12):e1000788. doi:10.1371/journal.pgen.1000788
80. Poirel L, Jayol A, Bontron S, et al. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother. 2014;70(1):75–80. doi:10.1093/jac/dku323
81. Gaibani P, Lombardo D, Lewis RE, et al. In vitro activity and post-antibiotic effects of colistin in combination with other antimicrobials against colistin-resistant KPC-producing Klebsiella pneumoniae bloodstream isolates. J Antimicrob Chemother. 2014;69(7):1856–1865. doi:10.1093/jac/dku065
82. Giani T, Arena F, Vaggelli G, et al. Large nosocomial outbreak of colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae by clonal expansion of an mgrB deletion mutant. J Clin Microbiol. 2015;53(10):3341–3344.
83. Wright MS, Suzuki Y, Jones MB, et al. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother. 2015;59(1):536–543. doi:10.1128/AAC.04037-14
84. Sherman EX, Hufnagel DA, Weiss DS. MCR-1 confers cross-resistance to lysozyme. Lancet Infect Dis. 2016;16(11):1226–1227. doi:10.1016/S1473-3099(16)30395-4
85. Hasman H, Hammerum AM, Hansen F, et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Eurosurveillance. 2015;20(49). doi:10.2807/1560-7917.ES.2015.20.49.30085
86. Perrin-Guyomard A, Bruneau M, Houée P, et al. Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Eurosurveillance. 2016;21(6):1–3. doi:10.2807/1560-7917.ES.2016.21.6.30135
87. Yao X, Doi Y, Zeng L, Lv L, Liu J-H. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 2016;16(3):288–289. doi:10.1016/S1473-3099(16)00057-8
88. Malhotra-Kumar S, Xavier BB, Das AJ, et al. Colistin-resistant Escherichia coli harbouring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect Dis. 2016;16(3):286–287. doi:10.1016/S1473-3099(16)00014-1
89. Zurfuh K, Poirel L, Nordmann P, Nüesch-Inderbinen M, Hächler H, Stephan R. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-β-lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother. 2016;60(4):2594–2595. doi:10.1128/AAC.00066-16
90. Quesada A, Ugarte-Ruiz M, Iglesias MR, et al. Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res Vet Sci. 2016;105:134–135. doi:10.1016/j.rvsc.2016.02.003
91. Battisti A. Antibiotic resistance—Italy: colistin, MCR-1, E. coli, turkeys, 2014. Available from: http://www.poultrymed.com/Poultrymed/Templates/showpage.asp?DBID=1&LNGID=1&TMID=178&FID=1868&PID=0&IID=30269. Accessed April 4, 2019.
92. Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis. 2016;16(3):293. doi:10.1016/S1473-3099(16)30197-9
93. Elnahriry SS, Khalifa HO, Soliman AM, et al. Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob Agents Chemother. 2016;60(5):3249–3250. doi:10.1128/AAC.00269-16
94. Doumith M, Godbole G, Ashton P, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. 2016;71(8):2300–2305. doi:10.1093/jac/dkw093
95. Rapoport M, Faccone D, Pasteran F, et al. First description of mcr-1-mediated colistin resistance in human infections caused by Escherichia coli in Latin America. Antimicrob Agents Chemother. 2016;60(7):4412–4413. doi:10.1128/AAC.00573-16
96. Yu CY, Ang GY, Chin PS, Ngeow YF, Yin W-F, Chan K-G. Emergence of mcr-1-mediated colistin resistance in Escherichia coli in Malaysia. Int J Antimicrob Agents. 2016;47(6):504. doi:10.1016/j.ijantimicag.2016.04.004
97. Falgenhauer L, Waezsada S-E, Yao Y, et al. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing gram-negative bacteria in Germany. Lancet Infect Dis. 2016;16(3):282–283. doi:10.1016/S1473-3099(16)00009-8
98. Perreten V, Strauss C, Collaud A, Gerber D. Colistin resistance gene mcr-1 in avian-pathogenic Escherichia coli in South Africa. Antimicrob Agents Chemother. 2016;60(7):4414–4415. doi:10.1128/AAC.00548-16
99. Poirel L, Nordmann P. Emerging plasmid-encoded colistin resistance: the animal world as the culprit? J Antimicrob Chemother. 2016;71(8):2326–2327. doi:10.1093/jac/dkw074
100. Haenni M, Poirel L, Kieffer N, et al. Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis. 2016;16(3):281–282. doi:10.1016/S1473-3099(16)00007-4
101. Du H, Chen L, Tang Y-W, Kreiswirth BN. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis. 2016;16(3):287–288. doi:10.1016/S1473-3099(16)00056-6
102. Yin W, Li H, Shen Y, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio. 2017;8(3):e00543–00517. doi:10.1128/mBio.00543-17
103. Carattoli A, Villa L, Feudi C, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance. 2017;22(31). doi:10.2807/1560-7917.ES.2017.22.31.30589
104. Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72(12):3317–3324. doi:10.1093/jac/dkx327
105. AbuOun M, Stubberfield EJ, Duggett NA, et al. mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother. 2017;72(10):2745–2749. doi:10.1093/jac/dkx286
106. Yang Y-Q, Li Y-X, Lei C-W, Zhang A-Y, Wang H-N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73(7):1791–1795. doi:10.1093/jac/dky111
107. Wang X, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122. doi:10.1038/s41426-018-0124-z
108. Kieffer N, Nordmann P, Poirel L. Moraxella species as potential sources of MCR-like polymyxin-resistance determinants. Antimicrob Agents Chemother. 2017;61(6):e00129-17.
109. Zurfluh K, Tasara T, Poirel L, Nordmann P, Stephan R. Draft genome sequence of Escherichia coli S51, a chicken isolate harboring a chromosomally encoded mcr-1 gene. Genome Announc. 2016;4(4):e00796–00716. doi:10.1128/genomeA.00796-16
110. De Majumdar S, Yu J, Fookes M, et al. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog. 2015;11(1):e1004627. doi:10.1371/journal.ppat.1004627
111. Raetz CR, Guan Z, Ingram BO, et al. Discovery of new biosynthetic pathways: the lipid A story. J Lipid Res. 2009;50(Supplement):S103–S108. doi:10.1194/jlr.R800060-JLR200
112. Llobet E, Tomás JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology. 2008;154(12):3877–3886. doi:10.1099/mic.0.2008/022301-0
113. Formosa C, Herold M, Vidaillac C, Duval R, Dague E. Unravelling of a mechanism of resistance to colistin in Klebsiella pneumoniae using atomic force microscopy. J Antimicrob Chemother. 2015;70(8):2261–2270. doi:10.1093/jac/dkv118
114. Lacour S, Doublet P, Obadia B, Cozzone AJ, Grangeasse C. A novel role for protein-tyrosine kinase Etk from Escherichia coli K-12 related to polymyxin resistance. Res Microbiol. 2006;157(7):637–641. doi:10.1016/j.resmic.2006.01.003
115. Lacour S, Bechet E, Cozzone AJ, Mijakovic I, Grangeasse C, Sonenshein AL. Tyrosine phosphorylation of the UDP-glucose dehydrogenase of Escherichia coli is at the crossroads of colanic acid synthesis and polymyxin resistance. PLoS One. 2008;3(8):e3053. doi:10.1371/journal.pone.0003053
116. Chambers JR, Sauer K. The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. J Bacteriol. 2013;195(20):4678–4688.
117. Srinivasan VB, Rajamohan G. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon is a SMR-type efflux pump involved in broad spectrum antimicrobial resistance. Antimicrob Agents Chemother. 2013;57(9):4449–4462.
118. Parra‐Lopez C, Baer MT, Groisman EA. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. Embo J. 1993;12(11):4053–4062.
119. Ni W, Li Y, Guan J, et al. Effects of efflux pump inhibitors on colistin resistance in multidrug-resistant Gram-negative bacteria. Antimicrob Agents Chemother. 2016;60(5):3215–3218. doi:10.1128/AAC.00248-16
120. Kallel H, Bahloul M, Hergafi L, et al. Colistin as a salvage therapy for nosocomial infections caused by multidrug-resistant bacteria in the ICU. Int J Antimicrob Agents. 2006;28(4):366–369. doi:10.1016/j.ijantimicag.2006.07.008
121. Batirel A, Balkan I, Karabay O, et al. Comparison of colistin–carbapenem, colistin–sulbactam, and colistin plus other antibacterial agents for the treatment of extremely drug-resistant Acinetobacter baumannii bloodstream infections. Eur J Clin Microbiol Infect Dis. 2014;33(8):1311–1322. doi:10.1007/s10096-014-2070-6
122. Petrosillo N, Chinello P, Proietti M, et al. Combined colistin and rifampicin therapy for carbapenem-resistant Acinetobacter baumannii infections: clinical outcome and adverse events. Clin Microbiol Infect. 2005;11(8):682–683. doi:10.1111/j.1469-0691.2005.01198.x
123. Saballs M, Pujol M, Tubau F, et al. Rifampicin/imipenem combination in the treatment of carbapenem-resistant Acinetobacter baumannii infections. J Antimicrob Chemother. 2006;58(3):697–700. doi:10.1093/jac/dkl274
124. Rodríguez-Hernández M-J, Pachón J, Pichardo C, et al. Imipenem, doxycycline and amikacin in monotherapy and in combination in Acinetobacter baumannii experimental pneumonia. J Antimicrob Chemother. 2000;45(4):493–501.
125. Sobieszczyk ME, Furuya EY, Hay CM, et al. Combination therapy with polymyxin B for the treatment of multidrug-resistant Gram-negative respiratory tract infections. J Antimicrob Chemother. 2004;54(2):566–569. doi:10.1093/jac/dkh369
126. Haddad F, Van Horn K, Carbonaro C, Aguero-Rosenfeld M, Wormser GJE, Diseases I. Evaluation of antibiotic combinations against multidrug-resistant Acinetobacter baumannii using the E-test. Eur J Clin Microbiol Infect Dis. 2005;24(8):577–579. doi:10.1007/s10096-005-1366-y
127. Ko W-C, Lee H-C, Chiang S-R, et al. In vitro and in vivo activity of meropenem and sulbactam against a multidrug-resistant Acinetobacter baumannii strain. J Antimicrob Chemother. 2004;53(2):393–395.
128. Dizbay M, Tozlu DK, Cirak MY, Isik Y, Ozdemir K, Arman DJT. In vitro synergistic activity of tigecycline and colistin against XDR-Acinetobacter baumannii. J Antibiot. 2010;63(2):51.
129. Jayol A, Poirel L, Brink A, et al. Resistance to colistin associated to a single amino acid change in protein PmrB among Klebsiella pneumoniae of worldwide origin. Antimicrob Agents Chemother. 2014;58(8):4762–4766.
130. Olaitan AO, Morand S, Rolain J-M. Emergence of Colistin-Resistant Bacteria in Humans without Colistin Usage: A New Worry and Cause for Vigilance. Int J Antimicrob Agents. 2015;45(6):600–604.
131. Kang KN, Klein DR, Kazi MI, et al. Colistin heteroresistance in Enterobacter cloacae is mediated by PmrAB-independent 4-amino-4-deoxy-l-arabinose addition to lipid A. bioRxiv. 2019;11:516872.
132. Johnson L, Horsman SR, Charron-Mazenod L, et al. Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiol. 2013;13(1):115.
133. Zeng K-J, Doi Y, Patil S, Huang X, Tian G-BJ. chemotherapy. Emergence of the plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob Agents Chemother. 2016;60(6):3862–3863. doi:10.1128/AAC.00345-16
134. Li X-P, Fang L-X, Jiang P, et al. Emergence of the colistin resistance gene mcr-1 in Citrobacter freundii. Int J Antimicrob Agents. 2017;49(6):786–787.
135. Garcia-Graells C, De Keersmaecker SC, Vanneste K, et al. Detection of plasmid-mediated colistin resistance, mcr-1 and mcr-2 genes, in Salmonella spp. Isolated from food at retail in Belgium from 2012 to 2015. Foodborne Pathog Dis. 2018;15(2):114–117. doi:10.1089/fpd.2017.2329
136. Litrup E, Kiil K, Hammerum AM, Roer L, Nielsen EM, Torpdahl MJE. Plasmid-borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009–17. Eurosurveillance. 2017;22(31).
137. Carretto E, Brovarone F, Nardini P, et al. Detection of mcr-4 positive Salmonella enterica serovar Typhimurium in clinical isolates of human origin, Italy, October to November 2016. Eurosurveillance. 2018;23(2):17–00821.
138. Chavda B, Lv J, Hou M, et al. Coidentification of mcr-4.3 and blaNDM-1 in a clinical Enterobacter cloacae isolate from China. Antimicrob Agents Chemother. 2018;62(10):e00649–00618. doi:10.1128/AAC.00649-18
139. Fukuda A, Sato T, Shinagawa M, et al. High prevalence of mcr-1, mcr-3 and mcr-5 in Escherichia coli derived from diseased pigs in Japan. Int J Antimicrob Agents. 2018;51(1):163–164. doi:10.1016/j.ijantimicag.2017.11.010
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
