Back to Journals » Infection and Drug Resistance » Volume 12
Increase in antibiotic resistant Helicobacter pylori in a University Hospital in Japan
Authors Kageyama C, Sato M, Sakae H, Obayashi Y, Kawahara Y, Mima T , Matsushita O, Yokota K, Mizuno M, Okada H
Received 29 November 2018
Accepted for publication 6 February 2019
Published 12 March 2019 Volume 2019:12 Pages 597—602
DOI https://doi.org/10.2147/IDR.S196452
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Chihiro Kageyama,1 Mayu Sato,1 Hiroyuki Sakae,2 Yuka Obayashi,2 Yoshiro Kawahara,2 Takehiko Mima,3 Osamu Matsushita,3 Kenji Yokota,1 Motowo Mizuno,4 Hiroyuki Okada2
1Graduate School of Health Science, Medical Technology, Okayama University, Okayama 700-8558, Japan; 2Gastroenterology and Hepatology, Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, Okayama 700-8558, Japan; 3Bacteriology, Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, Okayama 700-8558, Japan; 4Department of Gastroenterology and Hepatology, Kurashiki Central Hospital, 710-8602, Japan
Background: Eradication effectively prevents Helicobacter pylori-associated diseases; however, H. pylori antibiotic resistance has increased throughout Japan and worldwide. This study aimed to assess rates of resistance to antibiotics; amoxicillin, clarithromycin and metronidazole in a University Hospital in Japan.
Materials and methods: H. pylori (208 strains) were isolated from patients at the Okayama University Hospital in Japan. The minimum inhibitory concentrations (MIC) were determined using the mean values of the E-test to determine the antimicrobial susceptibilities of the strains. Sequencing and gene analysis were performed to analyze resistance genes to clarithromycin and amoxicillin.
Results: Rates of amoxicillin, clarithromycin, and metronidazole resistance were 13%, 48%, and 49%, respectively. Genetic analysis indicated that the A2143G point mutation in 23S rDNA is closely associated with the MIC of clarithromycin. The MIC in amoxicillin-resistant strains increased with an increase in the number of PBP1A amino acids mutations.
Conclusion: Genetic analysis for resistant strains is not clinically effective in cases of amoxicillin resistance. Numerous bacteria with already high antibiotic resistance rates have been isolated in large hospitals such as a University Hospital. For effective eradication therapy, MIC measurement should be considered via several methods.
Keywords: Helicobacter pylori, resistance, clarithromycin, amoxicillin, University Hospital, genotype
Introduction
Helicobacter pylori is a Gram-negative spiral bacterium usually found in the gastric mucosa. H. pylori is associated, not only with gut diseases such as, peptic ulcers, gastric mucosa-associated lymphoid tissue (gastric-MALT) lymphoma, and gastric cancer,1–4 but also with systemic diseases.5,6 Eradication therapy is effective to prevent diseases and is widely practiced nationwide in hospitals and clinics within Japan.7,8 In particular, eradication therapy for H. pylori has evidently reduced the incidence of gastric cancer. Antibiotic resistance in H. pylori has recently increased.9,10 There are many reports that antibiotic resistance in H. pylori has increased alarmingly worldwide, thereby greatly affecting therapeutic efficacy.11–15 Many difficult-to-diagnose patients or those wherein eradication therapy failed at other common hospitals are admitted at Okayama University hospital and are carefully treated because the hospital administers third-line treatment. Therefore, it is possible that more resistant bacteria may be isolated at Okayama University Hospital than at other general hospitals.
The resistant strain surveillance committee in the Japanese Society for Helicobacter research reported that the prevalence rates of clarithromycin and metronidazole resistance are 38.5% and 3.0%, respectively, among patients requiring first-line triple therapy with a proton pump inhibitor, amoxicillin, and clarithromycin.16 However, clarithromycin and metronidazole resistance rates, after unsuccessful eradication by first- or second-line therapy, increased up to 90.5% and 52.4%, respectively. Furthermore, the committee has been admonished owing to the drastic increase in clarithromycin-resistant strains in Japan. We performed antibiotic susceptibility tests to amoxicillin, clarithromycin, and metronidazole in isolated H. pylori strains in the Okayama University Hospital. SNPs involving substitution of adenine to guanine residues at positions 2142 and 2143 of the 23S rRNA of H. pylori (A2142G and A2143G) are reportedly associated with clarithromycin resistance.17 Amoxicillin resistance depends on mutations in the amino acid sequence of PBP1A.18
Therefore, we assessed antibiotic resistance rates in H. pylori strains to amoxicillin, clarithromycin, and metronidazole at the Okayama University Hospital. In addition, we performed a genotype analysis of clarithromycin resistance and mutations in PBP1A in amoxicillin-resistant H. pylori because these antibiotics are being used for not only first-line treatment but also for all treatments.
Materials and methods
Patients and materials
Gastric biopsy specimens were obtained with written informed consent from patients who underwent endoscopy at Okayama University Hospital from January 2011 to February 2016. Ethical approval to carry out the study was obtained a priori from the Okayama University Ethics Committee. Gastric biopsy specimens were obtained from the antrum and body for H. pylori culture. Biopsy samples were homogenized and spread onto both non-selective brain heart infusion (BHI) agar supplemented with 7% (v/v) sterile defibrinated horse blood and selective BHI horse blood agar containing trimethoprim (50 mg mL-1), vancomycin (100 mg mL-1), and polymyxin B (25,000 IU), and incubated at 37°C under micro-aerobic conditions for 7 days. H. pylori growth was monitored by assessing colony morphology and positive biochemical reactions, such as those with urease. H. pylori (208 strains) were isolated from 131 culture-positive patients. Cultured H. pylori strains were maintained in –80°C until use.
Antibiotic susceptibility tests
H. pylori susceptibility to clarithromycin and amoxicillin was quantified using the E-test.19,20 After initial bacterial isolation from biopsy samples, colonies were harvested from pure H. pylori cultures. The pure-cultured strain was sub-cultured cultured for 7 days and suspended in 1 mL sterile saline at McFarland 1.0. The suspension was spread on brain heart infusion (BHI) agar supplemented with 7% (v/v) horse blood, with sterile cotton swabs. E-test strips were placed on the plates in accordance with the manufacturer’s instructions. These plates were incubated at 37°C under micro-aerobic conditions for 7 days. Strains were considered resistant to amoxicillin, clarithromycin, and metronidazole if the MIC was ≧0.1 µg/mL, ≧2 µg/mL, and ≧8 µg/mL, respectively.
PCR amplification of the clarithromycin resistance gene
Bacterial DNA was extracted and purified using the QIAamp DNA Micro Kit (QIAGEN, Venlo, Netherlands). The most common point mutations (A2142G and A2143G) in 23S rDNA, conferring resistance to clarithromycin in H. pylori strains, were detected via PCR, using primers 23 S-F and 23 S-R (Table 1). The reaction mixture comprised 5 µL of 10× Ex Taq Buffer, 4 µL of dNTP Mixture, 1 µL of each primer, 1 µL of DNA template, 0.5 µL of TaKaRa Ex Taq (5-unit µl-1) and 38 µL of sterile distilled water. Cycling conditions were as follows: 98°C for 5 minutes, 35 cycles of denaturation at 98°C for 15 seconds, annealing at 60.5°C for 20 seconds, extension at 72°C for 1 minute, and final extension step at 72°C for 10 minutes. Five microliters of the PCR product were electrophoresed on a 2% agarose gel for 30 minutes and stained with ethidium bromide for 20 minutes, and bands were visualized using Dolphin-View (Dolphin-View, KURABO, Japan), an electrophoresis gel photographing apparatus.
  | Table 1 Primers for PCR |
PCR amplification of the amoxicillin resistance gene
Bacterial DNA was extracted and purified using QIAamp DNA Micro Kit (QIAGEN, Venlo, Netherlands). Mutations in PBP1A, conferring amoxicillin resistance in H. pylori strains, were also detected via PCR, using primers PBP1-F and PBP1-R (Table 1). The reaction mixture comprised 5 µL of 10× Ex Taq Buffer, 4 µL of dNTP Mixture, 1 µL of each primer, 1 µL of DNA template, 0.5 µL of TaKaRa Ex Taq (5-unit µl-1) and 38 µL of sterile distilled water. Cycling conditions were as follows: 35 cycles of denaturation at 94°C for 1 minute, annealing at 60°C for 1 minute, and extension at 72°C for 1 minute. PCR products were assessed via electrophoresis on a 2% agarose gel, which was photographed using Dolphin-View (Dolphin-View, KURABO, Japan).
Sequencing of PCR products
PCR-amplified DNA samples were purified via ethanol precipitation. The samples were sequenced using forward and reverse primers, using a DNA sequencer (PRISM3130 Genetic Analyzer, Applied Biosystems) at Hokkaido System Science Co., Ltd. (Sapporo, Japan). Sequences were analyzed using Genetyx software (https://genetyx.software.informer.com).
Statistical analysis
Statistical analysis of mutations in the clarithromycin resistance gene among resistant strains and susceptible strains was performed using the chi-squared test. Mutations in the amoxicillin resistance gene were analyzed using Bartlett’s test. P<0.05 was considered statistically significant.
Results
Resistance rate of H. pylori at the Okayama University Hospital
From January 2011 to February 2016, 208 strains of H. pylori were isolated from 131 culture-positive patients who underwent gastric biopsy at the Okayama University Hospital. These strains were tested for amoxicillin, clarithromycin, and metronidazole susceptibility via the E-test. The amoxicillin-resistant strains accounted for 26 of the 207 (13%) strains isolated; 100 of 208 (48%) were clarithromycin-resistant strains; and 101 of the 189 (49%) strains. Cumulative curves of MICs of these three antibiotics are shown in Figure 1.
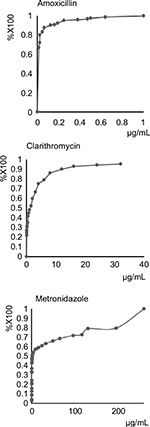  | Figure 1 Cumulative curves for the minimum inhibitory concentrations of amoxicillin, clarithromycin, and metronidazole. |
Clarithromycin resistance gene and resistance value
Mutations in the clarithromycin resistance gene (the most common point mutations being A2142G and A2143G in 23S rDNA) were examined via sequencing of 86 of the 208 (41%) strains. Thirty-seven of these isolates were resistant to clarithromycin (MIC≧2 mg/L), 27 displayed intermediate resistance (0.1≦MIC <2 mg/L), and 22 were sensitive (MIC <0.1 mg/L). Only two isolates harbored the A2142G mutation, which was not associated with clarithromycin resistance. The A2143G mutation was present in 81% of clarithromycin-intermediate-resistant isolates and in 95% of clarithromycin-resistant isolates (Table 2). The A2143G mutation is closely associated with the corresponding MIC values.
Amoxicillin resistance gene and resistance value
After sequencing of PBP1A from each isolated strain, amino acid mutations were detected at 14 points in the amoxicillin-resistant (AmxR) strains and 19 points in amoxicillin-sensitive (AmxS) strains. All mutations were not significantly associated with amoxicillin resistance (Table 3). These strains were categorized into 3 groups based on the MICs of amoxicillin: MIC <0.016, 0.016< MIC < 0.1, and 0.1< MIC. The amino acid mutation numbers were compared in each group. In the group with the lowest MICs (MIC <0.016, n=29), the average value for the mutation number was 2.14; intermediate MICs (0.016< MIC < 0.1, n=20), 2.35; highest MICs (0.1< MIC, n=8), 3.38. There was significant (P<0.05) difference between the lowest MIC and highest MIC groups (Figure 2). The number of mutations contributes to a higher MIC in AmxR strains.
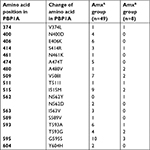  | Table 3 Amino acid substitutions in PBP1A in amoxicillin-sensitive and amoxicillin-resistant Helicobacter pylori strains Abbreviation: Amx, amoxicillin. |
Discussion
Antibiotic resistance of H. pylori is increasing; however, the resistance rate is still low during first-line therapy in Japan.21,22 Clarithromycin- and metronidazole-resistant strains have different geographical distributions worldwide and are divided into four groups. The US, Austria, and Japan constitute one group wherein clarithromycin resistance is high; however, metronidazole resistance is low.23 The prevalence rate of amoxicillin-resistant H. pylori is less than 1% in many areas.24–26 However, in the present study performed at Okayama University Hospital, the resistance rate of clarithromycin and metronidazole were high. In addition, the amoxicillin resistance rate was also high. In the near future, we believe that not only the clarithromycin and metronidazole resistance rates, but also the amoxicillin resistance rate will increase in Japan.
Clarithromycin resistance is associated with a mutation in 23S rDNA in the 50S ribosome. Mutations at both locations, ie, 2142 and 2143, involve an adenine-to-guanine transition, the latter leading to an adenine-to-cytosine trans-version at position 2142.27,28 The predominant mutations among resistant strains in Europe were A2143G (44.1%) and A2142G (32.6%). A recent study from India reported that the dominant point mutation was at the A2142G site, while the A2143G mutation was not detected.29 The prevalence of the A2143G as a primary mutation is 10–14% among the Chinese population; however, administration of clarithromycin to obliterate an H. pylori infection increased the prevalence (32%) of the A2143G mutation in Chinese patients.30 These reports indicate that the A2143G mutation in 23S mRNA is most important for clarithromycin resistance. In the present study, A2143G was associated with clarithromycin resistance. Other Japanese studies have also reported that A2143G was the primary mutation in clarithromycin-resistant strains.31–33 These mutations in 23S rDNA associated with clarithromycin resistance differed among different countries; therefore, genetic tests consistent with the strains prevalent in each country are required.
Amoxicillin resistance is caused by a mutation in PBP1A. Previous Japanese studies have reported that an amino acid substitution of Asn562→Tyr (N562Y) was similarly observed in all amoxicillin-resistant strains.34 Other substitutions including Ser414→Arg (S414R), Thr593→Ala (T593A), Gly595→Ser (G595S), and Ala599→Thr (A599T) were detected in amoxicillin-resistant strains in studies reported in Korea.35 The substitution of Ser414→Arg (S414R) has also been reported from Netherlands.36 Another study from Korea reported that the T593 substitution particularly contributes to amoxicillin resistance.12 However, in the present study, these substitutions were not detected or were detected in both resistant and sensitive strains. Similarly, no amino acid substitutions were detected in all eight amoxicillin-resistant strains. All detected amino acid substitutions were not specific for amoxicillin-resistant strains. The present results indicate that amoxicillin resistance in H. pylori is associated with multiple amino acid substitutions in PBP1A and tends to increase with an increase in the number of amino acid substitutions in PBP1A. Hence, it is better to avoid determining amoxicillin resistance exclusively on the basis of amino acid substitutions at a specific position. Currently, the detailed mechanism underlying amoxicillin resistance in H. pylori is unclear. Amoxicillin-resistant H. pylori in Japan are expected to increase in the future, and further studies on the acquisition of amoxicillin resistance are necessary.
Conclusion
Many bacteria with high antibiotic resistance have been isolated in large hospitals such as the Okayama University Hospital in Japan. To detect antibiotic resistance in H. pylori, culture-based assessment and SNP analysis are useful to detect clarithromycin-resistant bacteria. However, genetic analysis of PBP1A is not applicable for detecting amoxicillin resistance owing to the need to analyze amino acid substitution mutations. The method for detecting antibiotic resistance should be selected appropriately in accordance with the type of antibiotic.
Abbreviations
Amx, amoxicillin; BHI, brain hart infusion; MIC, minimum inhibitory concentration; PCR, polymerase chain reaction; SNP, Single Nucleotide Polymorphism.
Acknowledgments
We would like to thank all participants for their participation in this study. This work received no specific grant from any funding agency.
Disclosure
The authors report no conflicts of interest in this work.
References
Infection with Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum. 1994;61:177–240. | ||
Blaser MJ. The bacteria behind ulcers. Sci Am. 1996;274(2):104–107. | ||
Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345(11):784–789. | ||
Graham DY. Helicobacter pylori infection is the primary cause of gastric cancer. J Gastroenterol. 2000;35(Suppl 12):90–97. | ||
Franchini M, Veneri D. Helicobacter pylori infection and immune thrombocytopenic purpura: an update. Helicobacter. 2004;9(4):342–346. | ||
Ayada K, Yokota K, Kobayashi K, Shoenfeld Y, Matsuura E, Oguma K. Chronic infections and atherosclerosis. Clin Rev Allergy Immunol. 2009;37(1):44–48. | ||
Asaka M, Satoh K, Sugano K, et al. Guidelines in the management of Helicobacter pylori infection in Japan. Helicobacter. 2001;6(3):177–186. | ||
Asaka M, Mabe K, Matsushima R, Tsuda M. Helicobacter pylori eradication to eliminate gastric cancer: the Japanese strategy. Gastroenterol Clin North Am. 2015;44(3):639–648. | ||
Lee YC, Chiang TH, Chou CK, et al. Association between Helicobacter pylori eradication and gastric cancer incidence: a systematic review and meta-analysis. Gastroenterology. 2016;150(5):1113–1124. | ||
Rokkas T, Rokka A, Portincasa P. A systematic review and meta-analysis of the role of Helicobacter pylori eradication in preventing gastric cancer. Ann Gastroenterol. 2017;30(4):414–423. | ||
Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization Regions. Gastroenterology. 2018;155(5):1372–1382. | ||
Bachir M, Allem R, Benejat L, et al. Molecular detection of mutations involved in Helicobacter pylori antibiotic resistance in Algeria. J Antimicrob Chemother. 2018;73(8):2034–2038. | ||
Gunnarsdottir AI, Gudjonsson H, Hardardottir H, Jonsdottir KD, Bjornsson ES. Antibiotic susceptibility of Helicobacter pylori in Iceland. Infect Dis. 2017;49(9):647–654. | ||
Dang BN, Graham DY. Helicobacter pylori infection and antibiotic resistance: a WHO high priority? Nat Rev Gastroenterol Hepatol. 2017;14(7):383–384. | ||
Abadi ATB. Resistance to clarithromycin and gastroenterologist’s persistence roles in nomination for Helicobacter pylori as high priority pathogen by World Health Organization. World J Gastroenterol. 2017;23(35):6379–6384. | ||
Hashinaga M. Antibiotic resistant surveillance of Helicobacter pylori in Japan. Jpn J Helicobacter Res. 2016;17:45–49. Japanese. | ||
Nakamura A, Furuta T, Shirai N, et al. Determination of mutations of the 23S rRNA gene of Helicobacter pylori by allele specific primer-polymerase chain reaction method. J Gastroenterol Hepatol. 2007;22(7):1057–1063. | ||
Kwon YH, Kim JY, Kim N1, et al. Specific mutations of penicillin-binding protein 1A in 77 clinically acquired amoxicillin-resistant Helicobacter pylori strains in comparison with 77 amoxicillin-susceptible strains. Helicobacter. 2017;22(6):e12437. | ||
Glupczynski Y, Labbé M, Hansen W, Crokaert F, Yourassowsky E. Evaluation of the E test for quantitative antimicrobial susceptibility testing of Helicobacter pylori. J Clin Microbiol. 1991;29(9):2072–2075. | ||
Cederbrant G, Kahlmeter G, Ljungh A. The E test for antimicrobial susceptibility testing of Helicobacter pylori. J Antimicrob Chemother. 1993;31(1):65–71. | ||
Tanabe H, Yoshino K, Ando K, et al. Vonoprazan-based triple therapy is non-inferior to susceptibility-guided proton pump inhibitor-based triple therapy for Helicobacter pylori eradication. Ann Clin Microbiol Antimicrob. 2018;17(1):17–29. | ||
Kuo YT, Liou JM, El-Omar EM, et al. Asian Pacific Alliance on Helicobacter and Microbiota. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2017;2:707–715. | ||
Ansari S, Yamaoka Y. Current understanding and management of Helicobacter pylori infection: an updated appraisal. F1000Res. 2018;7:7211. | ||
Poon SK, Chang CS, Su J, et al. Primary resistance to antibiotics and its clinical impact on the efficacy of Helicobacter pylori lansoprazole-based triple therapies. Aliment Pharmacol Ther. 2002;16(2):291–296. | ||
Almeida N, Romãozinho JM, Donato MM, et al. Helicobacter pylori antimicrobial resistance rates in the central region of Portugal. Clin Microbiol Infect. 2014;20(11):1127–1133. | ||
Bachir M, Allem R, Tifrit A, et al. Primary antibiotic resistance and its relationship with cagA and vacA genes in Helicobacter pylori isolates from Algerian patients. Braz J Microbiol. 2018;49(3):544–551. | ||
Alarcón T, Vega AE, Domingo D, Martínez MJ, López-Brea M. Clarithromycin resistance among Helicobacter pylori strains isolated from children: prevalence and study of mechanism of resistance by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 2003;41(1):486–499. | ||
van Doorn LJ, Glupczynski Y, Kusters JG, et al. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob Agents Chemother. 2001;45(5):1500–1504. | ||
Ubhayawardana NL, Weerasekera MM, Weerasekera D, Samarasinghe K, Gunasekera CP, Fernando N. Detection of clarithromycin-resistant Helicobacter pylori strains in a dyspeptic patient population in Sri Lanka by polymerase chain reaction-restriction fragment length polymorphism. Indian J Med Microbiol. 2015;33(3):374–377. | ||
Liu Z1, Shen J, Zhang L, et al. Prevalence of A2143G mutation of H. pylori-23S rRNA in Chinese subjects with and without clarithromycin use history. BMC Microbiol. 2008; 8(8):81. | ||
Furuta T, Soya Y, Sugimoto M, et al. Modified allele-specific primer-polymerase chain reaction method for analysis of susceptibility of Helicobacter pylori strains to clarithromycin. J Gastroenterol Hepatol. 2007;22(11):1810–1815. | ||
Matsumoto H, Shiotani A, Katsumata R, et al. Helicobacter pylori eradication with proton pump inhibitors or potassium-competitive acid blockers: the effect of clarithromycin resistance. Dig Dis Sci. 2016;61(11):3215–3220. | ||
Miftahussurur M, Syam AF, Nusi IA, et al. Surveillance of Helicobacter pylori antibiotic susceptibility in indonesia: different resistance types among regions and with novel genetic mutations. PLoS One. 2016;11(12):e0166199. | ||
Rimbara E, Noguchi N, Kawai T, Sasatsu M. Correlation between substitutions in penicillin-binding protein 1 and amoxicillin resistance in Helicobacter pylori. Microbiol Immunol. 2007;51(10):939–944. | ||
Kim BJ, Kim JG. Substitutions in penicillin-binding protein 1 in amoxicillin-resistant Helicobacter pylori strains isolated from Korean patients. Gut Liver. 2013;7(6):655–660. | ||
Gerrits MM, Schuijffel D, van Zwet AA, Kuipers EJ, Vandenbroucke-Grauls CM, Kusters JG. Alterations in penicillin-binding protein 1A confer resistance to beta-lactam antibiotics in Helicobacter pylori. Antimicrob Agents Chemother. 2002;46(7):2229–2233. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


