Back to Journals » Infection and Drug Resistance » Volume 12
Prevalence and antimicrobial resistance of Shigella species isolated from diarrheal patients in Ahvaz, southwest Iran
Authors Sheikh AF, Moosavian M, Abdi M, Heidary M, Shahi F , Jomezadeh N , Seyed-Mohammadi S , Saki M, khoshnood S
Received 21 September 2018
Accepted for publication 5 December 2018
Published 22 January 2019 Volume 2019:12 Pages 249—253
DOI https://doi.org/10.2147/IDR.S187861
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Ahmad Farajzadeh Sheikh,1,2 Mojtaba Moosavian,1,2 Mahtab Abdi,2 Mohsen Heidary,3,4 Fatemeh Shahi,2 Nabi Jomehzadeh,5 Sakineh Seyed-Mohammadi,1,2,6 Morteza Saki,1,2,6 Saeed Khoshnood1,2,6
1Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 2Department of Microbiology, Faculty of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 3Department of Microbiology, Faculty of Medicine, Iran University of Medical Sciences,Tehran, Iran; 4Student Research Committee, School of Allied Medical Sciences, Iran University of Medical Sciences, Tehran, Iran; 5Abadan School of Medical Sciences, Abadan, Iran; 6Student Research Committee, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
Introduction: Shigellosis is a significant global human health problem, and Shigella is in charge of almost 165 million cases of this disease annually, of whom 163 million cases are in developing countries and 1.5 million cases are in developed countries. The main aims of the current survey were to identify Shigella spp. isolated from diarrheal patients by conventional biochemical tests, determine the antimicrobial susceptibility profiles by disk diffusion method, and detect the ipaH gene using the PCR assay.
Methods: The bacterial isolates were identified as Shigella spp. by microbiological tests and were serogrouped by the slide agglutination test. Antimicrobial susceptibility testing was performed using the disk diffusion method. PCR was performed to detect the ipaH gene.
Results: The Shigella strains were isolated from 522 patients with various diarrhea, including bloody diarrhea (3%), mucoid plus bloody diarrhea (1.9%), mucoid diarrhea (3.2%), and watery diarrhea (3.2%). Overall, 69 (13.2%) isolates were positive for Shigella spp., of which 34 (49.3%) serotypes were identified as Shigella flexneri, 22 (31.9%) serotypes were identified as Shigella sonnei, 9 (13%) serotypes were identified as Shigella boydii, and 4 (5.8%) serotypes were identified as Shigella dysenteriae. Antibiotic susceptibility tests revealed that the highest resistance percentage was related to ampicillin (82%) and trimethoprim-sulfamethoxazole (77%), and ciprofloxacin and ceftriaxone were the best antibiotics against Shigella isolates.
Conclusion: We concluded that Shigella spp. can be considered as an etiological agent of diarrhea in southwest Iran. Since the drug resistance pattern of Shigella differs geographically and over time within a country, continuous and regular surveillance program is necessary.
Keywords: Shigella, diarrhea, resistance, Iran
Introduction
The Shigella species are facultative intracellular gram-negative pathogens that cause shigellosis, which remains a significant public health concern. Shigella spp. was adopted as a genus of the family Enterobacteriaceae in the 1950s and serogrouped into the following four species: subgroup A (Shigella dysenteriae), subgroup B (Shigella flexneri), subgroup C (Shigella boydii), and subgroup D (Shigella sonnei).1,2
These enteric bacteria are in charge of almost 165 million cases of diarrhea annually, of whom 163 million cases are in developing countries and 1.5 million cases are in developed countries.3,4 The most common symptoms of shigellosis are light watery diarrhea, vomiting, fever, tenesmus, headache, and abdominal pain.
The illness is generally self-limiting but may become life threatening in patients with compromised immune systems or in the absence of adequate health care.5 Due to the low infectious dose of Shigella spp. (10–100 organisms) compared with other enteric pathogens, shigellosis is a serious public health threat.2 Lack of proper access to food sources and poor health care contribute to a high risk of morbidity and mortality in many developing countries. A variety of raw vegetables, salads, meat, milk, and other dairy products can serve as vehicles for the transmission of Shigella spp. Therefore, the most common causes of contamination are unsanitary practices of food handlers and fecally contaminated water.3
All age groups are affected by Shigella, but the age group under 5 years is most susceptible, because of low personal cleanliness and partially developed immunity and absence of past exposures.6 The proper and effective treatment for the disease leads to reduce the shedding of the bacteria and prevent lethal outcomes.
However, because of the antibiotic resistance development over the past half-century, the options for antimicrobial therapy in shigellosis are limited to a small number of drugs.7 Over time, the antimicrobial resistance patterns of Shigella spp. have changed according to geographical locations and the treatment process became more complicated.8
The main aims of this study were to identify Shigella spp. isolated from diarrheal patients by conventional biochemical tests, determine the antimicrobial susceptibility profiles by disk diffusion method, and detect the ipaH gene using the PCR assay.
Methods
Ethics statement
This study was approved by the ethics committee of Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran (No. IR.AJUMS.REC.1396.568). As a part of the Ahvaz Jundishapur University of Medical Sciences policy, written informed consent was obtained from all of the children’s parents or legal guardian of any patient under the age of 18 years. The study was conducted in accordance with the Declaration of Helsinki.
Study area and specimen collection
This cross-sectional study was performed in cooperation with Golestan teaching hospital at the Jundishapur University belonging to the Iranian health system in Ahvaz, Iran, during November 2016 to October 2017. Golestan hospital is the second largest university-affiliated hospital located in Ahvaz city (the capital of Khozestan state). Ahvaz city is located 820 km southwest of Tehran, the capital city of Iran. Stool samples were obtained from 522 patients aged 2–65 years who were distinguished as suffering from diarrhea by the clinical physician.
Diarrhea means watery or loose stools, usually at least three times in 24 hours. Mucus or blood can appear in the stools with some infections. The patients undergoing antibiotic therapy at the time of sample collection and those with persistence diarrhea were not included. The specimens were collected in sterile plastic containers and immediately transported to the Department of Microbiology of the Ahvaz Jundishapur University of Medical Sciences for processing.
Biochemical test
The fecal specimens were cultivated on selective and differential media, including MacConkey agar, xylose lysine desoxycholate (XLD) agar, and Salmonella–Shigella (SS) agar (EMD Millipore, Billerica, MA, USA), and then were incubated at 37°C for 24 hours. Identification of Shigella was carried out by subjecting presumptive colonies onto triple sugar iron (TSI) agar, methyl red (MR) broth, Voges–Proskauer (VP) broth, lysine iron agar (LIA), urea broth, indole test, sulfide indole motility (SIM) test, and citrate utilization tests, and incubated for 24–48 hours at 37°C.9,10
Serotyping
The bacterial isolates that were identified as Shigella by their morphological and biochemical features were emulsified in normal saline and mixed with an equal volume of specific Shigella antiserum (Baharafshan, Tehran, Iran). A macroscopic agglutination was considered as a positive reaction.11
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the Kirby–Bauer disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) recommendations.12 The susceptibilities of all isolates to different antibiotics were determined using ceftriaxone (CRO, 30 µg), erythromycin (ERY; 15 μg), ampicillin (AMP, 10 µg), chloramphenicol (CAM, 30 µg), nalidixic acid (NA, 30 µg), trimethoprim-sulfamethoxazole (SXT, 25 µg), ceftazidime (CAZ, 30 µg), ciprofloxacin (CIP, 5 µg), cefixime (CFM, 5 µg), and gentamicin, (GEN, 10 µg) (Mast Diagnostics Ltd., Merseyside, UK). Escherichia coli ATCC 25922 was used as quality control.
DNA extraction and PCR assay
DNA was extracted from Shigella colonies grown overnight on nutrient agar (EMD Millipore) by the boiling method. Briefly, bacteria were cultured in 5 mL of trypticase soy broth (EMD Millipore) and incubated for 24 hours at 37°C. Then, 200 µL of broth culture was mixed with 800 µL of sterile distilled water and the suspension was heated at 95°C for 10 minutes.
Then, the solution was centrifuged at 12,000× g for 5 minutes to remove any cell debris. Finally, 200 µL of the supernatant was stored at –20°C and used as the template for subsequent amplification. PCR amplification was performed to detect the ipaH gene in Shigella isolates.13 Amplification of the ipaH gene was carried out using a thermal gradient cycler (Eppendorf Co., Hamburg, Germany) with the following protocol14,15: the PCR mixture contained 2.5 mL of 10× buffer (10 mM Tris-HCl and 50 mM KCl), 1.5 mM MgCl2, 3 µL of DNA template, 200 µM each dNTPs, 0.4 µM of each forward and reverse ipaH primer, 0.75 U of Taq polymerase, and sterilized distilled water to complete the reaction volume (25 µL).
The expected sizes of PCR amplicons were revealed by electrophoresis on 1.5% horizontal agarose gel in Tris-borate-EDTA (TBE) buffer and stained with ethidium bromide (0.5 µg/mL) (SinaClon, Tehran, Iran). Primer sequences, PCR conditions, and the PCR product size are shown in Table 1.16
  | Table 1 Primers and PCR conditions for the amplification of ipaH gene |
Data analysis
The data were entered and analyzed using the SPSS software, Version 22.0 (IBM Corporation, Armonk, NY, USA) and Microsoft Excel 2016 (Microsoft Corporation, Redmond, WA, USA) statistical software.
Results
Stool samples were obtained from 522 diarrheal patients, and the mean age of patients was 30±1 years, with a range of 2–65 years. According to stool observation and clinical finding, 16 (3%) patients had bloody diarrhea, 26 (4.9%) patients had mucoid as well as bloody diarrhea, 10 (1.9%) patients had mucoid diarrhea, and 17 (3.2%) patients had watery diarrhea. Various age groups were ≤6 years (55%), 7–10 years (21.6%), 11–20 years (14.5%), and ≥21 years (8.8%). Three hundred and eighteen (60.9%) patients were male, and 204 (39.1%) patients were female (Table 2).
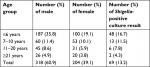  | Table 2 Description of groups of patients with positive result for Shigella culture in this study |
The patients have had various clinical symptoms, including headache (N=322, 61.7%), vomiting (N=141, 27%), fever (N=384, 73.5%), and abdominal pain (N=412, 78.9%). All the isolates were recognized by culture and serological tests, and then confirmed by the PCR method. In this study, 24 people were excluded from the study due to the use of antibiotics and 13 people were diagnosed with persistent diarrhea.
Overall, 69 (13.2%) isolates were confirmed as Shigella spp., of which 34 (49.2%) serotypes were identified as S. flexneri, 22 (31.9%) serotypes were identified as S. sonnei, 9 (13%) serotypes were identified as S. boydii, and 4 (5.8%) serotypes were identified as S. dysenteriae. All 69 isolates produced PCR amplicons of the ipaH gene of 500 bp.
Antibiotic susceptibility tests using the Kirby–Bauer method revealed that the highest resistance percentage was related to AMP (57, 82%), and SXT, 53 (77%), followed by NA (48, 69%), and ERY (47, 68.1%). In addition, Shigella spp. were resistant to the cephalosporins, of which 38 (55%) isolates were resistant to CFM and 33 (48%) isolates were resistant to CAZ.
The results showed that CIP and CRO were the best antibiotics against Shigella isolates, while more than 50% of S. dysenteriae strains were susceptible to NA, GEN, CRO, and CIP. Among the S. boydii strains, 55.5% were resistant to AMP, CFM, CAZ, and SXT. All S. dysenteriae strains were resistant to AMP, erythromycin, and SXT. Antibiotic resistance patterns of the four Shigella stereotypes are detailed in Table 3.
Discussion
Shigellosis is a significant global human health problem, and a more common serious condition in developing countries.3,4 Antibiotic therapy decreases the severity and duration of the infection and the fecal elimination of the organism, which in turn would prevent its further spread.17
The current study provides results of molecular characterization and antimicrobial resistance of Shigella spp. isolated from diarrheal patients in Iran. In this study, shigellosis was seen in all age groups and patients under the age of 10 years were the highest, because children in this age are most sensitive to shigellosis primarily due to the higher exposure to the contaminated environment, poor sanitation, and personal hygiene and less effective immune responses to Shigella spp.18
Identification of Shigella spp. as enteric pathogens with a universal impact has increased during recent years. In our study, Shigella spp. were isolated from 13.2% of diarrheal patients using cultural and biochemical methods, and the amplification of the ipaH gene has confirmed this isolation rate. This rate of isolation of Shigella was similar to studies in Iran (14%) and Brazil (10%) and, however, was different from studies in India (4%) and Ethiopia (4%).19–22 Furthermore, in our study, male and female children were equally affected, which is similar to the previous studies.20,23
This investigation showed that most of the Shigella isolates were resistant to AMP, SXT, and NA (up to 60%). In addition, Shigella spp. were resistant to the CFM (59.5%), CAZ (55%), and CRO (43.6%). Among antibiotics tested for the susceptibility of Shigella spp. isolates, CIP was the best with 76.6% sensitivity followed by CRO with 44% sensitivity.
In a study performed by Jafari et al,24 more than 90% of Shigella isolates were susceptible to CRO, CAZ, CFM, and CIP. In another study conducted by Mostafavi et al,25 Shigella serotypes have a very high level resistance to SXT and AMP and high level resistance to third-generation cephalosporins (>90% and 50% respectively).
All investigations performed in Iran during the last 20 years revealed a high level of resistance to SXT and AMP. Less than 30% of the Shigella isolates were sensitive to either of these two agents in Zahedan, Mashhad, and Zanjan provinces.6,25,26 The most common serotype isolated in our study was S. Flexneri, which was similar to Jomezadeh et al’s report in Iran. According to their report, S. flexneri (52.7%) was the most common serogroup and the most resistance was seen regarding the SXT (80.5%) and AMP (63.8%).8
Overall, the low resistance rate to CIP in our study showed that fluoroquinolones are still ideal drugs for treating the shigellosis in our region. Due to the limitation of fluoroquinolones’ prescription in children because of their side effects, the third-generation cephalosporins are used as a substitute treatment of shigellosis.25
Conclusion
We conclude that Shigella spp. can be considered as an etiological agent of diarrhea with a high level of antibiotic resistance in Ahvaz, Iran. Also, since the drug resistance pattern of Shigella differs in geographical regions and over time within a country, continuous and regular surveillance programs are necessary.
Acknowledgments
We would like to thank the Department of Microbiology, Faculty of Medicine, Ahvaz Jundishapur University of Medical Sciences, for their cooperation. Our appreciation goes to the Vice Chancellor for Research affairs, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran, and Tropical and Infectious Diseases Research Center of the University for their financial (Grant No. OG-96125) and executive support.
Disclosure
The authors report no conflicts of interest in this work.
References
Yang F, Yang J, Zhang X, et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33(19):6445–6458. | ||
Grimont F, Lejay-Collin M, Talukder KA, et al. Identification of a group of Shigella-like isolates as Shigella boydii 20. J Med Microbiol. 2007;56(Pt 6):749–754. | ||
Alipour M, Talebjannat M, Nabiuni M. Polymerase chain reaction method for the rapid detection of virulent Shigella spp. Int J Mol Clin Microbiol. 2012;2(1):134–137. | ||
Humphries RM, Linscott AJ. Laboratory diagnosis of bacterial gastroenteritis. Clin Microbiol Rev. 2015;28(1):3–31. | ||
Schroeder GN, Hilbi H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev. 2008;21(1):134–156. | ||
Gebrekidan A, Dejene TA, Kahsay G, Wasihun AG. Prevalence and antimicrobial susceptibility patterns of Shigella among acute diarrheal outpatients in Mekelle hospital, Northern Ethiopia. BMC Res Notes. 2015;8(1):611. | ||
Walker JC, Verma NK. Identification of a putative pathogenicity island in Shigella flexneri using subtractive hybridisation of the S. flexneri and Escherichia coli genomes. FEMS Microbiol Lett. 2002;213(2):257–264. | ||
Jomezadeh N, Babamoradi S, Kalantar E, Javaherizadeh H. Isolation and antibiotic susceptibility of Shigella species from stool samples among hospitalized children in Abadan, Iran. Gastroenterol Hepatol Bed Bench. 2014;7(4):218. | ||
Taneja N, Mewara A, Kumar A, Verma G, Sharma M. Cephalosporin-resistant Shigella flexneri over 9 years (2001-09) in India. J Antimicrob Chemother. 2012;67(6):1347–1353. | ||
Heidary M, Bahramian A, Hashemi A, et al. Detection of acrA, acrB, aac(6’)-Ib-cr, and qepA genes among clinical isolates of Escherichia coli and Klebsiella pneumoniae. Acta Microbiol Immunol Hung. 2017;64(1):63–69. | ||
Van der Ploeg CA, Viñas MR, Terragno R, Bruno SB, Binsztein N. Laboratory Protocol: Serotyping of Shigella spp. Geneva: WHO Global Foodborne Infections Network; 2010. | ||
Wayne P. Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Disk Diffusion Susceptibility Tests 19th ed. Approved Standard; CLSI document M100-S19; 2009. | ||
Sedighi M, Halajzadeh M, Ramazanzadeh R, Amirmozafari N, Heidary M, Pirouzi S. Molecular detection of β-lactamase and integron genes in clinical strains of Klebsiella pneumoniae by multiplex polymerase chain reaction. Rev Soc Bras Med Trop. 2017;50(3):321–328. | ||
Heidary M, Salimi Chirani A, Khoshnood S, et al. Molecular detection of aminoglycoside-modifying enzyme genes in Acinetobacter baumannii clinical isolates. Acta Microbiol Immunol Hung. 2017;64(2):143–150. | ||
Khoshnood S, Eslami G, Hashemi A. Distribution of aminoglycoside resistance genes among Acinetobacter baumannii strains isolated from burn patients in Tehran, Iran. Arch Pediatr Infect Dis. 2017;5(3):e57263. | ||
Na-Ubol M, Samosornsuk S, von Seidlein L, et al. Molecular characteristics of Shigella spp. isolated from patients with diarrhoea in a new industrialized area of Thailand. Epidemiol Infect. 2006;134(5):997–1003. | ||
Talebreza A, Memariani M, Memariani H, Shirazi MH, Shamsabad PE, Bakhtiari M. Prevalence and antibiotic susceptibility of Shigella species isolated from pediatric patients in Tehran. Arch Pediatr Infect Dis. 2016;4(1):e32395. | ||
Orrett FA. Prevalence of Shigella serogroups and their antimicrobial resistance patterns in southern Trinidad. J Health Popul Nutr. 2008;26(4):456–462. | ||
Nunes MR, Magalhães PP, Penna FJ, Nunes JM, Mendes EN. Diarrhea associated with Shigella in children and susceptibility to antimicrobials. J Pediatr. 2012;88(2):125–128. | ||
Demissie TA, Wubie T, Yehuala FM, Fetene M, Gudeta A. Prevalence and antimicrobial susceptibility patterns of Shigella and Salmonella species among patients with diarrhea attending Gondar Town Health Institutions, Northwest Ethiopia. Sci J Pub Health. 2014;2(5):469–475. | ||
Savadkoohi RB, Ahmadpour-Kacho M. Prevalence of Shigella species and their antimicrobial resistance patterns at Amirkola children hospital, North of Iran. Iran J Pediatr. 2007;17(2):118–122. | ||
Hosseini Nave H, Mansouri S, Sadeghi A, Moradi M. Molecular diagnosis and anti-microbial resistance patterns among Shigella spp. isolated from patients with diarrhea. Gastroenterol Hepatol Bed Bench. 2016;9(3):205. | ||
Dallal MMS, Eghbal M, Sharafianpour A, Zolfaghari MR, Yazdi MKS. Prevalence and multiple drug resistance of Shigella sonnei isolated from diarrheal stool of children. J Med Bacteriol. 2015;4(3–4):24–29. | ||
Jafari F, Hamidian M, Salmanzadeh-Ahrabi S. Molecular diagnosis and antimicrobial resistance pattern of Shigella spp. isolated from patients with acute diarrhea in Tehran, Iran. Gastroenterol Hepatol Bed Bench. 2009;1(1):11–17. | ||
Mostafavi N, Bighamian M, Mobasherizade S, Kelishadi R. Resistance of Shigella strains to extended-spectrum cephalosporins in Isfahan province. Med J Islam Repub Iran. 2016;30:428. | ||
Jamshidi A, Matbooei A. Shigella spp frequency, serotyping and antibiotic resistance pattern in acute diarrheic patients in Zanjan Shahid Beheshti Hospital, during 2003-2007. Zums J. 2008;16(62):77–84. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

