Back to Journals » International Journal of Nanomedicine » Volume 11
Apoptotic neuron-secreted HN12 inhibits cell apoptosis in Hirschsprung's disease
Authors Du C, Xie H, Zang R, Shen Z, Li H, Chen P, Xu X, Xia Y, Tang W
Received 12 June 2016
Accepted for publication 3 August 2016
Published 7 November 2016 Volume 2016:11 Pages 5871—5881
DOI https://doi.org/10.2147/IJN.S114838
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Linlin Sun
Chunxia Du,1,2,* Hua Xie,1,2,* Rujin Zang,1,2,* Ziyang Shen,1,2 Hongxing Li,1,2 Pingfa Chen,1,2 Xiaoqun Xu,1,2 Yankai Xia,2,3 Weibing Tang1,2
1Department of Pediatric Surgery, Nanjing Children’s Hospital Affiliated to Nanjing Medical University, 2State Key Laboratory of Reproductive Medicine, Institute of Toxicology, School of Public Health, 3Key Laboratory of Modern Toxicology, Ministry of Education, Nanjing Medical University, Nanjing, People’s Republic of China
*These authors contributed equally to this work
Abstract: Perturbation in apoptosis can lead to Hirschsprung’s disease (HSCR), which is a genetic disorder of neural crest development. It is believed that long noncoding RNAs (lncRNAs) play a role in the progression of HSCR. This study shows that apoptotic neurons can suppress apoptosis of nonapoptotic cells by secreting exosomes that contain high levels of HN12 lncRNA. Elevated exogenous HN12 in nonapoptotic cells effectively inhibited cell apoptosis by maintaining the function of mitochondria, including the production of ATP and the release of cytochrome C. These results demonstrate that secreted lncRNAs may serve as signaling molecules mediating intercellular communication in HSCR. In addition, high HN12 levels in the circulation worked as a biomarker for predicting HSCR, providing a potential, novel, noninvasive diagnostic approach for early screening of HSCR.
Keywords: Hirschsprung’s disease, neuronal development, exosomal long noncoding RNA, intercellular communication, apoptosis, mitochondria
Introduction
Hirschsprung’s disease (HSCR) is the main genetic cause of functional intestinal obstruction, with an incidence of one in 5,000 live births. This developmental disorder is associated with the absence of intrinsic ganglion cells along a variable length of the intestine.1 Although cell death does not play a major role during normal enteric nervous system (ENS) development, it may contribute to the etiology of HSCR.2 Apoptosis occurs normally during development, and works as a homeostatic mechanism to maintain cell populations in tissues. Proper neural crest-cell apoptosis contributes to a functional ENS.3 To date, research has indicated that the mitochondrial pathway is one of the two main apoptotic pathways.4 Long noncoding RNAs (lncRNAs) play an important role in various biological processes. For HSCR, microarray-expression profiling of dysregulated lncRNAs reveals their potential role in molecular diagnosis.5
Exosomes, nanosize microvesicles (30–100 nm in diameter), are released by both normal and diseased cells.6 After fusion with the plasma membrane, exosomes are secreted into extracellular space. These vesicles segregate the cargoes (lipids, proteins, and nucleic acids) within the membrane-covered vesicles.7–9 Over the past few years, evidences that microRNAs (miRNAs) can be secreted by cancer cells and transported to other cells via exosomes have accumulated.10–12 Interestingly, recent observations have also identified a vesicle-mediated transfer of lncRNAs as an important mechanism in the development of carcinoma.13,14
Humanin was originally identified as a 24-amino acid peptide that suppresses Alzheimer’s disease-related neuronal cell death, involving several distinct mechanisms.15 It was shown to restore cellular ATP levels and to protect neuronal cells by means of modulation of oxidative stress and apoptosis.16 It can be secreted from cells and can be found in plasma. There are 28 nuclear sequences throughout the human genome, and only 13 of them could generate functional peptides, including the MTRNR2L12 gene. HN12 is encoded by the MTRNR2L12 gene, and is identified as an lncRNA.17 It is classified as a pseudogene without possibility of protein coding in PubMed. Although recent study has revealed that HN12 can work as a candidate blood marker of early dementia in individuals with Down’s syndrome (DS),18 the mechanisms that regulate HN12 release and the potential biological functions of HN12 are completely unknown. As the presence of the MTRNR2L12 peptide has been confirmed in brain tissue and accumulated evidences have shown that ncRNA plays an important role in the pathogenesis of HSCR, we wanted to evaluate the potential role of HN12 in HSCR, especially in working as a candidate marker for HSCR.
HSCR occurs as an isolated phenotype in 70% of cases, but between 5% and 32% of patients have other associated congenital abnormalities. A large number of chromosomal anomalies have been described in HSCR patients. Free trisomy 21 (DS) is by far the most frequent, involving 2%–10% of cases.19,20 Association between HSCR and DS suggests that genetic factors that predispose to DS may be involved as an HSCR-susceptibility locus.
In this study, we demonstrate that HN12 is highly expressed in apoptosis-induced cells and can be released by secretive exosomes, which in turn are able to influence neighboring cells by protecting mitochondria and suppressing their apoptosis. Furthermore, our results suggest that HN12 lncRNA can be detected in serum and may serve as a biomarker for HSCR.
Materials and methods
Study population and sample recruitment
All experiments on human subjects were approved by the Institutional Ethics Committee of Nanjing Medical University (NJMU Birth Cohort), and all subjects gave written informed consent. These experiments were carried out in accordance with standard operating procedures. Total HSCR colon tissues, including the aganglionic zone and the matched distended region, that had been immediately frozen and stored at −80°C after surgery were recruited from the Department of Pediatric Surgery, Nanjing Children’s Affiliated Hospital between 2011 and 2014. The primary diagnosis was confirmed after barium enema and anorectal manometry evaluation. Eventual diagnosis of the HSCR was proved via pathological analysis for the aganglionosis. Negative controls were randomly picked out from patients who received surgical treatment because of intussusceptions or incarcerated and strangulated inguinal hernia without the ischemia or necrosis parts, but these patients were without HSCR or other congenital malformation. All subjects were Han Chinese.
Cell culture, transfection
SH-SY5Y (SY5Y) cells were cultured in complete growth Dulbecco’s Modified Eagle’s Medium (HyClone; GE Healthcare, Little Chalfont, UK), supplemented with 10% heat-inactivated fetal bovine serum (10%), penicillin (100 U/mL), and streptomycin (100 μg/mL) at 37°C, 5% CO2. The small interfering RNA (siRNA) against HN12 and negative controls (Table S1) was purchased from RealGene SRL (Reggio Calabria, Italy). Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used in all of the transfection experiments following the manufacturer’s instructions.
Cell-death assay and cell-apoptosis assay
The SY5Y cells were exposed to H2O2 to induce cell death. Different concentrations of H2O2 were added to cell cultures with or without fetal bovine serum for 24 hours, and then cell apoptosis was measured according to the manufacturer’s instructions using an annexin V–fluorescein isothiocyanate (FITC)/propidium iodide kit (KeyGen Biotech, Nanjing, People’s Republic of China). Apoptosis rates were analyzed using a flow cytometer (FACSCalibur; BD Biosciences, San Jose, CA, USA).
Morphological assessment of apoptosis
SY5Y cells were plated in a confocal plate. After 24 hours, cells were incubated with H2O2 for 24 hours, then washed with phosphate-buffered saline (PBS) twice prior to Hoechst 33342 (10 μg/mL) addition, and then incubated in the dark for 20 minutes. Morphologic change was observed with the laser confocal fluorescence microscopy.
Immunofluorescence
The cells were fixed in 4% paraformaldehyde, washed, and then permeabilized with 0.25% Triton X-100. Anti-TOMM20 antibody (ab78547; Abcam, Cambridge, UK) was used to stain mitochondria. The secondary antibody was FITC-labeled goat antirabbit IgG from Beyotime (A0562; Nantong, People’s Republic of China). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole; Beyotime). Images were visualized under a 100× oil objective using confocal microscopy. Quantification of images was conducted with Image-Pro Plus software.
Exosome purification from cell-culture supernatants
For exosomes secreted by culture cell lines, the culture medium was collected and cleared by centrifugation at 500× g for 15 minutes and then at 12,500× g for 20 minutes at 4°C. Exosomes were isolated by ultracentrifugation at 110,000× g for 70 minutes and washed in PBS using the same ultracentrifugation conditions. When indicated, 1 μM DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate; Beyotime) was added to the PBS and incubated for 20 minutes before a further round of PBS washing. The pelleted exosomes were resuspended in ~50 μL of PBS and subjected to cell treatment, Western blot analysis, or RNA extraction by Trizol LS (Thermo Fisher Scientific). For cell treatment, exosomes from 107 cells were added to 106 cells.
RNA isolation and quantitative real-time PCR
Total RNA, containing lncRNA, was extracted from tissue specimens and cell lines by using Trizol reagent (Life Technologies, CA, USA) according to the manufacturer’s instructions. We employed quantitative real-time polymerase chain reaction (qRT-PCR) to detect the expression levels of RNA. Human GAPDH RNA was used as a control for the detection of RNA. LncRNA or mRNA levels were calculated according to 2−ΔΔCt. Forward (F) and reverse (R) primer sequences are shown in Table S1.
Evaluation of level of intracellular ATP
The ATP assay was from Beyotime, and ATP was measured by luminometric methods using commercially available luciferin/luciferase reagents according to manufacturers’ instructions. The relative ATP level was calculated by ATP value/protein value. The protein value of the sample was measured by the bicinchoninic acid method (Beyotime).
Protein extraction and Western blotting
Total proteins were extracted from cultured cells or purified exosomes using radioimmunoprecipitation-assay buffer containing protease inhibitors, while isolation of mitochondrial and cytosolic proteins was carried out using a mitochondria/cytosol fractionation kit (Beyotime). Protein concentrations were measured by the bicinchoninic acid method (Beyotime). Western blot analysis was performed using standard procedures. Primary antibodies, including anti-COXIV, anti-cytochrome C, and anti-GAPDH antibody, were purchased from Beyotime, as well as the secondary antibodies, including HRP-linked antirabbit and HRP-linked antimouse. The anti-CD63 antibody was from Abcam.
Statistical methods
All experiments were repeated independently in triplicate at least. Differences between two independent groups were tested with Student’s t-test. Experimental data of tissue samples are presented as box plots of medians and range of log-transformed relative expression level using Wilcoxon rank-sum (Mann–Whitney) tests, and data are expressed as mean ± standard error of mean. Statistical analysis was performed by Stata 9.2, and presented with GraphPad Prism software. Results were considered to be statistically significant at P<0.05.
Results
Clinical information analysis
Clinical information, including age, sex, and body weight, was obtained from participants among 48 HSCR patients and 48 normal controls. The ages of HSCR patients and matched controls were 127.5±8.81 and 118.43±8.42 days, while body weights were 5.54±0.17 and 5.32±0.17 kg, respectively. None of the clinical information showed any significant differences between HSCR cases and normal controls (Table 1).
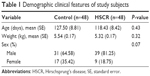  | Table 1 Demographic clinical features of study subjects |
HN12 inhibits cell apoptosis by protecting mitochondria
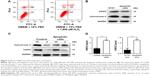
To begin with, HN12 expression in 48 paired HSCR samples and control samples was examined. HN12 was overexpressed in HSCR, as shown in Figure 1A, indicating HN12 might be involved in the pathological development of HSCR disease. Treatment with HN12 siRNA promoted cell apoptosis in SY5Y cells (Figure 1B), but had no effect on cell proliferation or migration (Figure S1A). It has been reported that HN blocks Bax translocation to mitochondria and suppresses cytochrome C release.21 Other research reported that HN induced an increase of ATP in both Bax-dependent and -independent mechanisms.22 To demonstrate whether HN12 works in the same way, siRNA against HN12 was treated with SY5Y cells. We found that HN12 reduced cytochrome C release from mitochondria (Figure 1C) and increased the level of ATP (Figure 1D). COXIV is required to drive ATP synthesis.23 Knockdown of HN12 also displayed lower COXIV expression (Figure 1E). These results revealed that knockdown of HN12 might promote cell apoptosis by destroying the function of the mitochondria.
Mitochondrial form and function are intimately linked.24 Altered mitochondrial dynamics have also been implicated in human diseases.25 The number of mitochondria was decreased when cells were treated with siRNA against HN12 (Figure 1F). A coactivator of nuclear receptors, PPARGC1A can stimulate mitochondria biogenesis and respiratory chain function, including the synthesis of ATP and expression of cytochrome C.26 Recent work has indicated that mitochondrial hyperfusion also serves to maintain ATP production.27 MFN1 and MFN2 have been shown to regulate mitochondrial fusion coordinately, and are essential for embryonic development.28 Therefore, expression levels of MFN1, MFN2, and PPARGC1A were detected in cells with siRNA against HN12 (Figure 1G). Then, relative expression levels of MFN1 and PPARGC1A were also measured in HSCR tissues (Figure S1B). Altered levels of MFN1 and PPARGC1A were determined to be the early stages that contributed to the apoptosis and the dysfunctional mitochondria with siRNA against HN12.
HN12 is highly expressed and can be secreted by apoptotic SY5Y cells
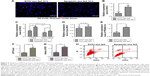
To evaluate the ectopic effect of HN12, SY5Y cells, chosen as apoptotic cell model, were treated with different doses of H2O2 (Figure S1C). SY5Y cells treated with 1,600 μmol/L H2O2 for 24 hours markedly induced SY5Y-cell apoptosis (Figure 2A). Examination of cell morphology using Hoechst staining is shown in Figure S1D. H2O2-induced cells presented with nuclear condensations compared with non-H2O2 treatment groups. To investigate whether mitochondria are involved in H2O2-induced apoptosis, the expression level of COXIV and the release of cytochrome C were detected in the apoptotic cells. As shown in Figure 2B, the protein levels of COXIV and cytochrome C were both decreased in apoptotic cells. Also, H2O2 induced the release of cytochrome C from mitochondria in apoptotic cells (Figure 2C). These results indicated that apoptotic cells were triggered by H2O2. Furthermore, impaired mitochondria were involved in the pathway of apoptosis, which was induced by H2O2.
The enrichment of HN12 in apoptotic SY5Y cells was confirmed using qRT-PCR (Figure 2D), which indicated that SY5Y cells exposed to H2O2 stimuli expressed increased levels of HN12. What is more, we explored whether HN12 was involved in cell–cell communication between apoptotic SY5Y cells and neighboring SY5Y cells. At first, in order to evaluate the ability of SY5Y cells and apoptotic cells to release exosomes, exosomes were collected by ultracentrifugation. The exosomes of SY5Y cells and apoptotic cells for HN12 secretion were then examined. The apoptotic cells secreted significantly elevated HN12 when compared with untreated SY5Y cells (Figure 2D). Overall, our data demonstrate that apoptotic SY5Y cells specifically secrete high levels of HN12 lncRNA into exosomes.
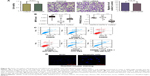
Apoptotic cell-secreted HN12 inhibits apoptosis of recipient cells
To investigate the biological functions of secreted HN12, exosomes labeled with the fluorescent dye DiI were incubated with untreated SY5Y cells. The recipient cells exhibited efficient uptake of exosomes regardless of the producer cells, as indicated by fluorescence microscopy (Figure 3A). In these cells, exosomes that were high in HN12 caused significantly increased intracellular HN12 (Figure 3B). Moreover, donor cells were treated with HN12 siRNA, then the exosomes of donor cells were isolated and incubated with recipient cells, showing that the increase of HN12 in recipient cells could be suppressed by treatment with siRNA against HN12 (Figure 3C). The pre-HN12 levels in recipient cells were unchanged by treatment with exosomes from either SY5Y cells or apoptotic cells (Figure 3D). These results indicated that the increase of HN12 in recipient SY5Y cells reflected the exosome-mediated HN12 transfer, but not an induction of endogenous expression. Increased expression of PPARGC1A was also observed in high-HN12 exosome-treated SY5Y cells, along with increased levels of ATP (Figure 3E and F). Also, mitochondria were upregulated in cells fed with exosomes derived from apoptotic cells (Figure 3G). MFN1 and MFN2 were not detected in those recipient cells. In addition, high-HN12 exosomes significantly prevented the apoptosis of recipient cells (Figure 3H).
HN12 acts as a biomarker for diagnosis of HSCR
lncRNAs have been reported as biomarkers for predicting survival, metastasis, and in the diagnosis of multiple diseases.29,30 HN12 was overexpressed in HSCR tissues and can be secreted by apoptotic cells, so we wondered whether HN12 could work as a biomarker for the diagnosis for HSCR. The expression level of HN12 was further examined by qRT-PCR in individual samples (21 cases and 14 controls). This showed that the expression of HN12 in HSCR plasma was significantly higher than in controls (Figure 4A). Receiver-operating characteristic (ROC)-curve analysis was then conducted to assess the diagnostic sensitivity and specificity of the lncRNA HN12 for HSCR. As presented in Figure 4B, the area under the curve of HN12 was 0.918.
Discussion
In the current study, we demonstrated that HN12 might contribute to the development of HSCR by apoptosis assay after confirming its high expression in HSCR clinical samples. What is more, our research is the first to discover that HN12 is highly secreted by apoptotic cells via exosomes and can inhibit recipient cells apoptosis by protecting mitochondria. Moreover, HN12 might work as molecular biomarker for diagnosis of HSCR.
Exosomes, which can deliver their content to target cells, are identified as biologically functional active signaling intermediates. In addition, several studies have described exosomes as signaling extracellular organelles that modulate the tumor microenvironment and promoting tumor progression.31,32 Many studies have also shown that the microenvironment influences the pathogenesis of HSCR.33,34 Our work is also the first time to identify the existence of exosomes in HSCR. Although more research is required to demonstrate the relationship between exosomes and the microenvironment in HSCR, it is supposed that exosomes possibly work through an abnormal extracellular matrix or through interactions with growth factors that are essential for intestinal neuronal network formation. In addition, exosomes have been considered as a novel platform for cancer therapy.35 Through the untiring efforts of researchers, we speculate that exosomes may work as potent therapeutics in HSCR, as it can deliver drugs to selective targets. Certainly, more studies are needed to apply exosomes in a drug-delivery system.
Humanin is particularly attractive due to its cytoprotective properties in the central nervous system.16 HN12, a humanin isoform, plays a possible role in the early development of dementia in DS. As DS is the most common chromosomal abnormality associated with HSCR, it may be suggested naturally that HN12 plays a role in HSCR. Our results showed that HN12 works in the same way as humanin. Its antiapoptotic function was shown to be due to the regulation of mitochondria, including the production of ATP and release of cytochrome C. What is more, mitochondrial number and function are altered in response to external stimuli in eukaryotoes.36 It has been reported that inhibition of mitochondria fission delays cytochrome C release.37 Our work further demonstrated that dysfunctional mitochondria were associated with the abnormal expression of MFN1 and PPARGC1A when HN12 was downregulated.
The involvement of lncRNA in HSCR is becoming increasingly recognized.38,39 LncRNAs are emerging as molecular players in several biological processes, acting at epigenetic, transcriptional, and posttranscriptional levels or processing small ncRNAs.40 However, a role for lncRNAs as intercellular signaling mediators has not been defined, and the potential of extracellular vesicles to transfer lncRNA is not well known. Our study identified a novel lncRNA gene in HN12 that is capable of functioning as an intercellular signaling mediator and modulating neuron behavior.
HSCR is the most common disorder of the ENS at birth. It is usually diagnosed by a barium enema, anorectal manometry, and a biopsy of the rectum. In other words, rich experience is required for correct diagnosis. Therefore, more reasonable and early screening strategies are needed for early HSCR diagnosis. Specific serum miRNA profiles in HSCR have revealed that miRNAs could be of considerable clinical value in the molecular diagnosis of HSCR. In this study, the level of HN12 in serum was measured and ROC-curve analyses then conducted. The lncRNA HN12 was considered a potential biomarker for HSCR in our study. However, our study had several limitations. With the poor stability of blood-based lncRNA, only 48 patient samples were analyzed, which limits study confidence. A larger sample size is needed to validate the diagnostic capability of HN12.
In summary, this work is the first to demonstrate that HN12 is highly expressed in HSCR and H2O2-induced apoptotic cells. It was found that HN12 that was induced in apoptotic cells could be packaged into exosomes, which were then transferred to possible target cells in order to protect recipient cells from apoptosis by providing enough functional mitochondria. Moreover, HN12 was identified to have the potential to be a predictive marker for HSCR.
Acknowledgments
The authors thank Dr Jie Zhang, Huan Chen, and Changgui Lu (Nanjing Children’s Hospital Affiliated to Nanjing Medical University) for sample collection. This study was supported by the Natural Science Foundation of China (NSFC 81370473, 81400574, 81570467), Natural Science Foundation of Jiangsu Province of China (BK20131388), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Disclosure
The authors report no conflicts of interest in this work.
References
Amiel J, Sproat-Emison E, Garcia-Barcelo M, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. | ||
Tjaden NE, Trainor PA. The developmental etiology and pathogenesis of Hirschsprung disease. Transl Res. 2013;162:1–15. | ||
McKeown SJ, Stamp L, Hao MM, Young HM. Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip Rev Dev Biol. 2013;2:113–129. | ||
Ouyang L, Shi Z, Zhao S, et al. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. | ||
Shen Z, Du C, Zang R, et al. Microarray expression profiling of dysregulated long non-coding RNAs in Hirschsprung’s disease reveals their potential role in molecular diagnosis. Neurogastroenterol Motil. 2016;28:266–273. | ||
Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–1494. | ||
Thoms M, Thomson E, Baßler J, Gnädig M, Griesel S, Hurt E. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell. 2015;162:1029–1038. | ||
Guo L, Guo N. Exosomes: potent regulators of tumor malignancy and potential bio-tools in clinical application. Crit Rev Oncol Hematol. 2015;95:346–358. | ||
Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. | ||
Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–144. | ||
Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. | ||
Singh R, Pochampally R, Watabe K, Lu Z, Mo YY. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol Cancer. 2014;13:256. | ||
Kogure T, Yan IK, Lin WL, Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer. 2013;4:261–272. | ||
Conigliaro A, Costa V, Lo Dico A, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. | ||
Yen K, Lee C, Mehta H, Cohen P. The emerging role of the mitochondrial-derived peptide humanin in stress resistance. J Mol Endocrinol. 2013;50:R11–R19. | ||
Hashimoto Y, Niikura T, Tajima H, et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Aβ. Proc Natl Acad Sci U S A. 2001;98:6336–6341. | ||
Bodzioch M, Lapicka-Bodzioch K, Zapala B, Kamysz W, Kiec-Wilk B, Dembinska-Kiec A. Evidence for potential functionality of nuclearly-encoded humanin isoforms. Genomics. 2009;94:247–256. | ||
Bik-Multanowski M, Pietrzyk JJ, Midro A. MTRNR2L12: a candidate blood marker of early Alzheimer’s disease-like dementia in adults with Down syndrome. J Alzheimers Dis. 2015;46:145–150. | ||
Jones KL, Pivnick EK, Hines-Dowell S, et al. A triple threat: Down syndrome, congenital central hypoventilation syndrome, and Hirschsprung disease. Pediatrics. 2012;130:e1382–e1384. | ||
Jannot AS, Pelet A, Henrion-Caude A, et al. Chromosome 21 scan in Down syndrome reveals DSCAM as a predisposing locus in Hirschsprung disease. PloS One. 2013;8:e62519. | ||
Guo B, Zhai D, Cabezas E, et al. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423:456–461. | ||
Kariya S, Hirano M, Furiya Y, Ueno S. Effect of humanin on decreased ATP levels of human lymphocytes harboring A3243G mutant mitochondrial DNA. Neuropeptides. 2005;39:97–101. | ||
Acin-Perez R, Gatti DL, Bai Y, Manfredi G. Protein phosphorylation and prevention of cytochrome oxidase inhibition by ATP: coupled mechanisms of energy metabolism regulation. Cell Metab. 2011;13:712–719. | ||
Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. | ||
Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. | ||
Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. | ||
Rolland SG, Motori E, Memar N, et al. Impaired complex IV activity in response to loss of LRPPRC function can be compensated by mitochondrial hyperfusion. Proc Natl Acad Sci U S A. 2013;110:E2967–E2976. | ||
Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. | ||
Pandey GK, Mitra S, Subhash S, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. | ||
Veltri WR. Non-coding RNAs as biomarkers for metastatic prostate cancer. Lancet Oncol. 2014;15:1412–1413. | ||
Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. | ||
Yuan A, Farber EL, Rapoport AL, et al. Transfer of microRNAs by embryonic stem cell microvesicles. PloS One. 2009;4:e4722. | ||
Barlow AJ, Dixon J, Dixon MJ, Trainor PA. Balancing neural crest cell intrinsic processes with those of the microenvironment in Tcof1 haploinsufficient mice enables complete enteric nervous system formation. Hum Mol Genet. 2012;21:1782–1793. | ||
Hagl CI, Rauch U, Klotz M, et al. The microenvironment in the Hirschsprung’s disease gut supports myenteric plexus growth. Int J Colorectal Dis. 2012;27:817–829. | ||
Tominaga N, Yoshioka Y, Ochiya T. A novel platform for cancer therapy using extracellular vesicles. Adv Drug Deliv Rev. 2015;95:50–55. | ||
Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G. Mitochondrial control of cellular life, stress, and death. Circ Res. 2012;111:1198–1207. | ||
Prudent J, Zunino R, Sugiura A, Mattie S, Shore GC, McBride HM. MAPL SUMOylation of Drp1 stabilizes an ER/mitochondrial platform required for cell death. Mol Cell. 2015;59:941–955. | ||
Xie H, Zhu D, Xu C, et al. Long none [sic] coding RNA HOTTIP/HOXA13 act as synergistic role by decreasing cell migration and proliferation in Hirschsprung disease. Biochem Biophys Res Commun. 2015;463:569–574. | ||
Luo Y, Li S, Teng Y, et al. Differential expression of FOXA1, DUSP6, and HA117 in colon segments of Hirschsprung’s disease. Int J Clin Exp Pathol. 2015;8:3979–3986. | ||
Mondal T, Subhash S, Vaid R, et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. |
Supplementary materials
Cell-proliferation assays
The CCK-8 assay (Beyotime, Nantong, People’s Republic of China) was used to detect cell proliferation. Briefly, 5,000 SH-SY5Y cells were seeded on 96-well plates. At indicated time points, 10 μL of the CCK-8 solution was added to each well, then the Infinite M200 multimode microplate reader (Tecan, Männedorf, Switzerland) was employed in measuring the absorbance at 450 nm. All experiments were performed in triplicate independently.
Cell transwell assays
About 100 μL cell suspension with serum-free medium was seeded in the upper chamber (106 cells/mL), and the lower chamber was filled with medium containing 10% fetal bovine serum. After 48-hour incubation at 37°C, nonmigratory cells in the upper chamber were removed by a cotton swab, and the cells that had migrated to the lower chamber were stained with crystal violet staining solution (Beyotime) and photographed under 40× magnification (five views per well). All experiments were performed in triplicate independently.
  | Table S1 Sequences of primers for quantitative real-time polymerase chain reaction and small interfering RNA-related sequence |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.