Back to Journals » Infection and Drug Resistance » Volume 11
Antiviral resistance markers in influenza virus sequences in Mexico, 2000–2017
Authors Toledo-Rueda W, Rosas-Murrieta NH , Muñoz-Medina JE , González-Bonilla CR, Reyes-Leyva J , Santos-López G
Received 3 October 2017
Accepted for publication 6 December 2017
Published 11 October 2018 Volume 2018:11 Pages 1751—1756
DOI https://doi.org/10.2147/IDR.S153154
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
William Toledo-Rueda,1,2 Nora H Rosas-Murrieta,3 José E Muñoz-Medina,4 César R González-Bonilla,4 Julio Reyes-Leyva,1 Gerardo Santos-López1
1Laboratory of Molecular Biology and Virology, Eastern Biomedical Research Center, Mexican Institute of Social Security, Metepec, Puebla, Mexico; 2Postgraduate in Chemical Sciences, Autonomous University of Puebla, Puebla, Mexico; 3Laboratory of Biochemistry and Molecular Biology, Chemistry Center, Institute of Science, Autonomous University of Puebla, Puebla, Mexico; 4Division of Laboratories for Surveillance and Epidemiological Research, Coordination of Epidemiological Surveillance, Mexican Institute of Social Security, Mexico City, Mexico.
Background: Influenza causes high rates of morbidity and mortality. Genetic variability of influenza viruses generates resistance to antivirals, which are of two types, since they act on two different viral targets: adamantanes, which block the M2 ion channel, and the neuraminidase (NA) inhibitors.
Methods: In Mexico, the available studies on the antiviral resistance of circulating influenza strains are scarce, so this work undertook an analysis of the Mexican sequences reported in public gene banks to perform a systematic analysis of the antiviral resistance markers on both M2 and NA. In all, 284 M2 sequences and 423 NA sequences were retrieved from three genetic databases (sequences from 2000 to 2017 were considered).
Results: The resistance markers to M2 blockers were present in 100% of H1N1 pdm2009, 83.6% of H3N2, and 5.8% of seasonal H1N1 sequences. Two resistance markers conferring resistance to NA inhibitors were present in seasonal H1N1 sequences, H275Y (50.0%) and N70S (33.3%). None of these viruses had both resistance markers, which are associated with oseltamivir resistance. The more frequent resistance marker in H1N1 pdm2009 NA sequences was H275Y, present in 3.6%, while S247N was present in 0.30%. Only one of the resistance-associated markers (Q136K) in NA (1.5%) was present in the analyzed H3N2 sequences, while sequences of influenza B virus did not present resistance markers to NA inhibitors. Some influenza A H1N1 pdm2009 sequences (1.8%) presented resistance markers to both M2 and NA.
Conclusion: Based on the present analysis, 7.1% of the all serotypes of influenza virus A sequences analyzed in Mexico from 2000 to 2017 have mutations conferring resistance to NA inhibitors. Because of this, and the limited availability of influenza drugs, it is necessary to increase the epidemiological surveillance, including molecular analysis, which will provide data such as the presence of changes associated with antiviral resistance.
Keywords: influenza A virus, drug resistance, M2 blockers, neuraminidase inhibitors, oseltamivir, zanamivir
Introduction
Influenza viruses belong to the family Orthomyxoviridae and are classified into three types: A, B, and C.1 Influenza A and B viruses are associated with high seasonal morbidity and mortality; however, influenza A is a special case because it occasionally causes pandemics and has more rapid evolution than types B and C.2,3
Influenza A and B viruses cause epidemics each year, associated with a variable number of deaths, absenteeism, hospitalization, and drug costs, as well as other public and private expenditures.4 To counteract influenza, vaccines and antivirals have been developed; however, in any of the cases, the protection and/or therapeutic capacity are not enough to eradicate or reduce, to the desired degree, the effects of infection in all patients.
The failure of vaccines and therapies is largely due to the genetic changes in influenza viruses, which are reflected in phenotypic variations that allow it to escape the host immune response, induce drug resistance, and/or cause pathogenic changes.5,6 In this regard, surveillance of anti-influenza drug resistance has great global relevance. There are two classes of drugs worldwide approved for treatment of illness caused by influenza virus, M2 ion channel blockers (such as amantadine and rimantadine) and neuraminidase (NA) inhibitors (such as zanamivir, oseltamivir, and peramivir).7
In recent years, most of the circulating viruses are resistant to adamantanes; therefore, they are not recommended for antiviral treatment. Hence, only NA inhibitors remain as the indicated anti-influenza drugs against the currently circulating viruses.7,8
In Mexico, the available studies on the drug resistance of circulating influenza strains are scarce, so this study undertook an analysis of sequences reported in public gene banks to perform a systematic analysis of the resistance molecular markers on both M2 and NA proteins.
Methods
Sequences of NA and M2 from influenza A and NA from influenza B viruses were retrieved from Influenza Research Database (www.fludb.org), Influenza Virus Database (https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=database), and GISAID EpiFlu (http://platform.gisaid.org/epi3/frontend). The inclusion criteria of the study were sequences of human influenza viruses reported from Mexico of any year, any subtype, or lineage, regardless of whether they are complete or partial. Sequences from 2000 to 2017 were considered. The redundant sequences and all sequences that did not contain the antiviral resistance positions were discarded from the analysis of those positions. Results were reported as sequences containing the specific marker with respect to the number of sequences analyzed on the related amino acid position. The amino acid positions related to resistance to M2 and NA inhibitors considered in the present study are summarized in Tables 1 and 2, respectively. The selected sequences were aligned using ClustalW Multiple Sequence Alignment tool (Clustal, Dublin, Ireland) in BioEdit v. 7.2.5 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).9
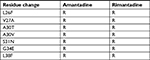  | Table 1 Mutations associated with adamantane resistance in influenza A viruses Note: Data from Belshe et al,23 Hay et al,24 and Abed et al.25 Abbreviation: R, resistance. |
Results
Overview of retrieved sequences
After discarding the redundant entries from gene bank entries, we undertook the study with 284 M2 and 423 NA sequences. All these sequences were subclassified as complete or partial, depending on whether or not they had the corresponding complete open reading frame.
M2 sequences
In this study, 284 M2 sequences were included, of which 199 (69%) were complete and 85 (31%) were partial: 205 (139 complete and 66 partial) were of H1N1 pdm2009, 17 (11 complete and 6 partial) of seasonal H1N1, 61 (48 complete and 13 partial) of H3N2, and 1 (complete) of H7N3. The M2 sequences were found between 2003 and 2017, and most of them were reported as of 2009 (70%); in fact, 2009 was the year in which the largest proportion of sequences were deposited (55%).
NA sequences
A total of 423 NA sequences were included. Of these, 197 (46%) were complete and 226 (54%) were partial: 420 corresponded to influenza A and 3 to influenza B. Of influenza A, 329 (134 complete and 195 partial) corresponded to H1N1 pdm2009, 70 (48 complete and 22 partial) to H3N2, 20 (13 complete and 7 partial) to seasonal H1N1, and 1 (complete) to H7N3. The 3 (1 complete and 2 partial) influenza B virus sequences corresponded 1 to Yamagata and 2 to Victoria linages. Sequences were found in the period 2000–2017, but, as in the case of M2, most of them are reported as of 2009 (77%), and specifically 57% corresponded to the 2009 pandemic.
Resistance to M2 blockers
The analyzed sequences showed a high frequency of the molecular marker S31N, which is associated with resistance to M2 blockers. All sequences (205/205) of H1N1 pdm2009 and 83.6% (51/61) of H3N2 viruses presented this marker. Seasonal H1N1 viruses had S31N in a much lower proportion: 5.8% (1/17) (Table 3). All other resistance markers were not found in the analyzed sequences. The overall resistance, including all sequences of influenza A viruses, was 90.5%.
  | Table 3 Analysis of adamantane resistance in influenza A virus sequences Abbreviation: RM, resistance marker. |
The most frequent codon for N31 resistance marker in H1N1 pdm09 and H3N2 viruses was AAU, which is present in 100% sequences bearing the resistance marker.
Resistance to NA inhibitors
The analysis of amino acid substitutions in NA related to resistance included those markers to well-known drugs oseltamivir and zanamivir and the recently approved drugs peramivir and laninamivir (Table 2).
Nine amino acid substitutions were analyzed in the H1N1 pdm2009 sequences, and only two changes were present in some of them. The most frequent was H275Y, present in 12/328 (3.6%), while S247N was present in 1/329 (0.30%). These changes and the rest of resistance markers are summarized in Table 4. All viruses carrying the marker H275Y had the codon UAC (12/12), while the other viruses had CAC (316/316). The only one virus carrying the marker S247N presented the codon AAU, and the other viruses had AGU (328/328). None of these viruses had both resistance markers, which are associated with oseltamivir resistance. The S247N change is also associated with zanamivir resistance and was the only sequence detected for this drug. No sequence had amino acids changes associated with peramivir and laninamivir resistance.
To analyze the resistance of seasonal H1N1 viruses, five amino acid substitutions were considered (Table 2). Only two resistance markers were present, H275Y in 10/20 (50.0%) and N70S in 6/18 (33.3%) sequences. None of these viruses had both resistance markers, which are associated with oseltamivir resistance (Table 5). No sequence had markers associated with zanamivir, peramivir, and laninamivir resistance. The most frequent codon in H274Y change was UAU (9/10), while the other viruses had CAC (7/10) and CAU (3/10). In all cases of N70S, the codon was AGC (6/6), and the other viruses presented AAC (12/12).
The analyses of NA resistance markers in H3N2 viruses included eleven positions (Table 2). Only one of the resistance-associated genotypic markers (Q136K) was present in the 70 (1.5%) analyzed N2 sequences. In the case of influenza B virus, none of the considered eight NA resistance markers was present in the three analyzed sequences. The overall rate of NA resistance markers, including all subtypes of influenza A viruses, was 7.1% (30/423).
Combined resistance
Some of viruses presented two or more resistance markers. H1N1 pdm2009 viruses with combined M2 and NA sequences (171) show that all of them were resistant to adamantanes and three (1.8%) were also resistant to oseltamivir.
Seasonal H1N1 and H3N2 viruses had 12 and 61 available sequences to analyze both genes, but none of them had double resistance, neither seasonal H1N1 nor H3N2 had resistance markers to zanamivir.
Discussion
Although vaccination as a form of prevention for influenza is widespread and relatively successful, there is still a high frequency of cases of influenza, which can lead to high severity and present a significant lethality. The NA inhibitors began to be used as specific drugs to reduce viral replication between 1999 and 2000 and have been widely recommended for treatment, especially in severe infections. However, the resistance presented by influenza viruses is significant and represents a global health concern.10
Currently, the NA inhibitors are the antiviral of choice for influenza treatment, although WHO estimates that 2% of circulating strains of influenza virus are resistant to these inhibitors, due to certain characteristic mutations causing of antiviral effectivity of drugs. The predominant resistance-associated change is H275Y (N1 numbering).11 Based on the present analysis, 7.1% of the sequences analyzed in Mexico have mutations conferring resistance to NA inhibitors (oseltamivir and zanamivir).
In Mexico, there are few studies reporting oseltamivir resistance in influenza viruses. These reports present values ranging from 0% to 0.33%.12–15
The percentage of resistant strains may fluctuate depending on the country and geographical area: in USA, it has been reported at 1.2%,16 in Brazil 1.4%,17 in Argentina 0.1%,18 and in Australia and Japan 18%.19,20 In the sequences analyzed in this work, the predominant mutation was H275Y in both the H1N1 pdm09 and seasonal H1N1 subtypes, while the H3N2 subtype did not present this mutation (H274Y), and its overall resistance proportion was lower than that of the other two influenza A viruses.
There is a gradual increase in circulating resistant strains worldwide since before 2008: the percentage of resistant strains was <1%21 and is currently 2% at global level. However, in some countries, high rates, >60%, have been reported.22
In the present analysis, the sequences of the H1N1 pdm09 during the period 2009–2012 present a higher frequency of H275Y, which could suggest that this mutation is being adapted, maintained, and disseminated in this subtype in Mexico. However, after 2012, none of analyzed NA sequences presented this resistance marker; therefore, it is difficult to predict the behavior of resistance in influenza viruses.
Because of this, and the limited availability of influenza drugs, it is necessary to increase the epidemiological surveillance, including molecular analysis, which will provide data such as the presence of changes associated with antiviral resistance, but can offer other phenotypic-associated features related to sequence changes, such as specificity for receptors, relationship with circulating strains, and antigenicity.
Conclusion
The clinical implications of resistance to antivirals should not be taken lightly. Based on the present analysis, 7.1% of circulating influenza viruses from 2000 to 2017 in Mexico have mutations that confer resistance to NA inhibitors. These resistant viruses increased their presence as of 2009; among them, H275Y (or H274Y) has been maintained and disseminated by different influenza virus subtypes around the world. This, coupled with therapeutic limitations against infection, forces us to establish better epidemiological surveillance systems that include the search for mutations that confer resistance to NA inhibitors, among other biological characteristics such as replication and transmissibility.
Acknowledgment
This work was supported by Funds for Scientific Infrastructure from the IMSS (CTFIS/10RD/12/2011) and FIS/IMSS/PROT/G09/775.
Disclosure
William Toledo-Rueda was supported by CONACYT (406868) and IMSS (98226171). Julio Reyes-Leyva has a research fellowship from Fundación IMSS A.C. The authors report no other conflicts of interest in this work.
References
Shaw ML, Palese P. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol 1. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2013. | ||
Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med. 2000;51:407–421. | ||
Wright PF, Neumann G, Kawaoka Y. Fields Virology. Vol 1. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2013. | ||
Peasah SK, Azziz-Baumgartner E, Breese J, Meltzer MI, Widdowson MA. Influenza cost and cost-effectiveness studies globally-a review. Vaccine. 2013;31(46):5339–5348. | ||
Belser JA, Maines TR, Tumpey TM, Katz JM. Influenza A virus transmission: contributing factors and clinical implications. Expert Rev Mol Med. 2010;12:e39. | ||
Medina RA, García-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9(8):590–603. | ||
Pizzorno A, Abed Y, Boivin G. Influenza drug resistance. Semin Respir Crit Care Med. 2011;32(4):409–422. | ||
Dong G, Peng C, Luo J, et al. Adamantane-resistant influenza a viruses in the world (1902–2013): frequency and distribution of M2 gene mutations. PLoS One. 2015;10(3):e0119115. | ||
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. | ||
Gubareva LV, Besselaar TG, Daniels RS, et al. Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2015-2016. Antiviral Res. 2017;146:12–20. | ||
Gubareva LV, Nedyalkova MS, Novikov DV, Murti KG, Hoffmann E, Hayden FG. A release-competent influenza A virus mutant lacking the coding capacity for the neuraminidase active site. J Gen Virol. 2002;83(Pt 11):2683–2692. | ||
Ramirez-Gonzalez JE, Gonzalez-Duran E, Alcantara-Perez P, et al. Oseltamivir-resistant pandemic (H1N1) 2009 virus, Mexico. Emerg Infect Dis. 2011;17(2):283–286. | ||
de La Rosa-Zamboni D, Vazquez-Perez JA, Avila-Rios S, et al. Molecular characterization of the predominant influenza A(H1N1)pdm09 virus in Mexico, December 2011-February 2012. PLoS One. 2012;7(11): e50116. | ||
Téllez-Sosa J, Rodríguez MH, Gómez-Barreto RE, et al. Using high-throughput sequencing to leverage surveillance of genetic diversity and oseltamivir resistance: a pilot study during the 2009 influenza A(H1N1) pandemic. PLoS One. 2013;8(7):e67010. | ||
Mosqueda-Gomez JL, Belaunzaran-Zamudio PF, Barba A, Cordova-Villalobos JA, Cuellar-Rodriguez JM, Ernesto Macias A. Surveillance of Oseltamivir-Resistant Influenza A(H1N1) pdm09 in Guanajuato State, Mexico from 2009 to 2012. Rev Invest Clin. 2015;67(4):235–239. | ||
Okomo-Adhiambo M, Fry AM, Su S, et al. Oseltamivir-resistant influenza A(H1N1)pdm09 viruses, United States, 2013-14. Emerg Infect Dis. 2015;21(1):136–141. | ||
Souza TM, Resende PC, Fintelman-Rodrigues N. Detection of oseltamivir-resistant pandemic influenza A(H1N1) pdm2009 in Brazil: can community transmission be ruled out? PLoS One. 2013;8(11):e80081. | ||
Pontoriero A, Avaro M, Benedetti E, et al. Surveillance of antiviral resistance markers in Argentina: detection of E119V neuraminidase mutation in a post-treatment immunocompromised patient. Mem Inst Oswaldo Cruz. 2016;111(12):745–749. | ||
Kiso M, Mitamura K, Sakai-Tagawa Y, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet. 2004;364(9436):759–765. | ||
Hurt AC, Hardie K, Wilson NJ, et al. Characteristics of a widespread community cluster of H275Y oseltamivir-resistant A(H1N1)pdm09 influenza in Australia. J Infect Dis. 2012;206(2):148–157. | ||
Sheu TG, Deyde VM, Okomo-Adhiambo M, et al. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother. 2008;52(9):3284–3292. | ||
Hurt AC, Holien JK, Parker MW, Barr IG. Oseltamivir resistance and the H274Y neuraminidase mutation in seasonal, pandemic and highly pathogenic influenza viruses. Drugs. 2009;69(18):2523–2531. | ||
Belshe RB, Smith MH, Hall CB, Betts R, Hay AJ. Genetic basis of resistance to rimantadine emerging during treatment of influenza virus infection. J Virol. 1988;62(5):1508–1512. | ||
Hay AJ, Zambon MC, Wolstenholme AJ, Skehel JJ, Smith MH. Molecular basis of resistance of influenza A viruses to amantadine. J Antimicrob Chemother. 1986;18(Suppl B):19–29. | ||
Abed Y, Goyette N, Boivin G. Generation and characterization of recombinant influenza A (H1N1) viruses harboring amantadine resistance mutations. Antimicrob Agents Chemother. 2005;49(2):556–559. | ||
Cheung CL, Rayner JM, Smith GJ, et al. Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. J Infect Dis. 2006;193(12):1626–1629. | ||
Wang MZ, Tai CY, Mendel DB. Mechanism by which mutations at his274 alter sensitivity of influenza a virus n1 neuraminidase to oseltamivir carboxylate and zanamivir. Antimicrob Agents Chemother. 2002;46(12):3809–3816. | ||
Little K, Leang SK, Butler J, et al. Zanamivir-resistant influenza viruses with Q136K or Q136R neuraminidase residue mutations can arise during MDCK cell culture creating challenges for antiviral susceptibility monitoring. Euro Surveill. 2015;20(45). | ||
Mckimm-Breschkin JL. Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance. Influenza Other Respir Viruses. 2013;7(Suppl 1):25–36. | ||
Le QM, Kiso M, Someya K, et al. Avian flu: isolation of drug-resistant H5N1 virus. Nature. 2005;437(7062):1108. | ||
Baz M, Abed Y, Mcdonald J, Boivin G. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin Infect Dis. 2006;43(12):1555–1561. | ||
Ison MG. Influenza in hospitalized adults: gaining insight into a significant problem. J Infect Dis. 2009;200(4):485–488. | ||
Nitsch-Osuch A, Brydak LB. Influenza viruses resistant to neuraminidase inhibitors. Acta Biochim Pol. 2014;61(3):505–508. | ||
Takayama I, Nakauchi M, Fujisaki S, Odagiri T, Tashiro M, Kageyama T. Rapid detection of the S247N neuraminidase mutation in influenza A(H1N1)pdm09 virus by one-step duplex RT-PCR assay. J Virol Methods. 2013;188(1–2):73–75. | ||
Yen HL, Hoffmann E, Taylor G, et al. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J Virol. 2006;80(17):8787–8795. | ||
Zürcher T, Yates PJ, Daly J, et al. Mutations conferring zanamivir resistance in human influenza virus N2 neuraminidases compromise virus fitness and are not stably maintained in vitro. J Antimicrob Chemother. 2006;58(4):723–732. | ||
Gubareva LV, Bethell R, Hart GJ, Murti KG, Penn CR, Webster RG. Characterization of mutants of influenza A virus selected with the neuraminidase inhibitor 4-guanidino-Neu5Ac2en. J Virol. 1996;70(3):1818–1827. | ||
Kamali A, Holodniy M. Influenza treatment and prophylaxis with neuraminidase inhibitors: a review. Infect Drug Resist. 2013;6:187–198. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



