Back to Journals » Infection and Drug Resistance » Volume 12
Antibacterial mechanism of peptide Cec4 against Acinetobacter baumannii
Authors Peng J, Long H, Liu W , Wu Z, Wang T, Zeng Z, Guo G, Wu J
Received 1 May 2019
Accepted for publication 11 July 2019
Published 5 August 2019 Volume 2019:12 Pages 2417—2428
DOI https://doi.org/10.2147/IDR.S214057
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Jian Peng,1–3 Huiling Long,3 Weiwei Liu,2,3 Zhaoying Wu,1,3 Tao Wang,3 Zhu Zeng,1–3 Guo Guo,2,3 Jianwei Wu3
1Key Laboratory of Biology and Medical Engineering, Department of Biotechnology, School of Biology & Engineering, Guizhou Medical University, Guiyang 550004, People’s Republic of China; 2Key Laboratory of Environmental Pollution Monitoring and Disease Control, Ministry of Education, Guizhou Medical University, Guiyang 550004, People’s Republic of China; 3Key and Characteristic Laboratory of Modern Pathogen Biology, Department of Human Parasitology, School of Basic Medical Sciences, Guizhou Medical University, Guiyang 550004, People’s Republic of China
Background: A case of Acinetobacter baumannii (A. baumannii), known as gram-negative bacteria, causes a range of nosocomial infections. Due to the continuous detection of multi-drug resistant A. baumannii in the clinic, there is an urgent need to find alternative therapies, including broad-spectrum antibacterial peptides (AMP). Recently it has been found that the peptide Cec4 has good antibacterial activity against A. baumannii, but the antibacterial mechanism remains elusive.
Materials and methods: The basic structure of Cec4 was analyzed by circular dichroism (CD) spectroscopy, and the potential antibacterial mechanism of Cec4 was detected by flow cytometry, transmission electron microscopy, fluorescence and confocal microscopy. The minimum inhibitory concentration (MIC) of antimicrobial peptides against various A. baumannii was determinated with broth microdilution techniques. The biofilm formation and the sensitivity detection of biofilms to antimicrobial peptides were detected by crystal violet staining.
Results: In this study, the main secondary structure of the antibacterial peptide Cec4 is α-helix (99.7%) in the hydrophobic environment. Furthermore, after the treatment with Cec4, an amount of leakage of A. baumannii and the destruction of its cell membrane were detected. Moreover, it was observed that FITC-Cec4 can enter the cell, and more cells were held in the G1 phase with peptide Cec4. However, the DNA binding assay of the peptide Cec4 indicates that the peptide does not target DNA. In addition, peptide Cec4 was superior in reducing adherent biofilms of A. baumannii compared to conventional antibiotics and has no cytotoxicity.
Conclusion: It is apparent that the antibacterial peptide Cec4 may achieve rapid sterilization by multi-target interaction and presents an attractive therapeutic option for the prevention and control of A. baumannii infections.
Keywords: antimicrobial peptides, A. baumannii, antibacterial mechanism, cell membrane
Introduction
A. baumannii is a major Gram-negative hospital pathogen and is often associated with severe hospital-acquired infections, including pneumonia, urinary tract infections, and bacteremia, especially in intensive care units.1,2 Due to the extensive use of antibiotics, more and more multi- and pan-resistant A. baumannii have been isolated in the hospital environment.3,4 These A. baumannii are resistant to several major classes of antibiotics, including the carbapenems known as the last line of defense, as well as tigecyclines and colistins.5,6 High-efficiency drug- resistant gene transfer ability, easy to produce drug inactivation enzyme, low permeability of the outer membrane and efficient efflux pump system are the main reasons leading to serious drug resistance of A. baumannii.7 More seriously, A. baumannii is easy to form biofilms on solid surfaces such as implants and medical devices, and the drug resistance of biofilm formed A. baumannii is 10–1000 times higher than that of planktonic bacteria.8 Therefore, the development of new antibiotics is urgently needed to solve the public health problems of antibiotic shortage and drug resistance.9
As an important component of innate immunity, antimicrobial peptides (AMPs) are small peptides with broad-spectrum antibacterial effects produced by the body after being stimulated by pathogens.10 They have better thermal stability than traditional antibiotics.11 Besides, they are considered the most promising alternative to antibiotics, as they do not easily cause bacterial resistance.12 At present, determination of the antibacterial mechanism of these peptides on bacteria is mainly concentrated on the bacterial cell membrane and intracellular activity.13,14 Studies have shown that Musca domestica cecropin (Mdc) can affect the cell membrane integrity of Escherichia coli and A. baumannii15 and also DNA binding to the E. coli bacterial genome in vitro.16 In addition, Xinjiang silkworm antibacterial peptide Cecropin-XJ can compete with Ethidium bromide (EB) for binding to Staphylococcus aureus DNA, and recombinant silkworm antibacterial peptide CM4 can bind to fungal DNA and RNA.17 Therefore, different antimicrobial peptides have different antibacterial mechanisms. However, one of the main reasons for the slow progress of the application of antimicrobial peptides is that most antimicrobial peptides have low antibacterial activity. For example, novel engineered peptide P307SQ-8C was an effective antimicrobial against multidrug-resistant A. baumannii with the MIC of 62.5 μg/mL,18 and a Cecropin A hybrid peptide CA (1–8) M (1–18) inhibits A. baumannii with the MIC of 45.7 μg/mL.19
Our previous study found that the peptide Cec4 has good antibacterial activity against A. baumannii with the MIC of 4 μg/mL, and its antibacterial effect is more durable than that of polymyxin B.20 In addition, the antibacterial activity is significantly better than Musca domestica cecropin antibacterial peptide Cec-1 (Mdc, Mdce) and three other highly homologous molecules.21,22 Therefore, in our study, the antibacterial mechanism of the peptide Cec4 against A. baumannii from the cell membrane and intracellular aspects was investigated. This will lay a theoretical foundation for the early development of a novel, highly active antibacterial agent.
Materials and methods
Bacterial isolates, peptides and reagents
1) A. baumannii (ATCC 19606) was preserved by the Key Laboratory of Modern Pathogenic Biology of Guizhou Medical University; Multidrug-resistant A. baumannii (MRAB) (ID: 4367661) and Extensive drug-resistant A. baumannii (PRAB) (ID: 4367992) were provided by the affiliated hospital of Guizhou Medical University. In addition, the Institutional Review Board (IRB) of Guizhou Medical University approves the collection and use of clinical strains. The strain ATCC 19606 of A. baumannii was used as the standard for comparison in biofilm formation and other assays.23 Strains were stored in glycerol, frozen at −80 °C, and cultured in Mueller-Hinton Broth (MHB) medium for 18–20 hrs at 37 °C before use.
2) The sequence of Cec4 presented in this article has been submitted to the GenBank database under accession number MG209110. Synthetic antibacterial peptide Cec4 (GWLKKIGKKIERVGQNTRDATIQAIGVAQQAANVAATLKG), Cec4-7 (GWLKKIGKKIERVGQHTRDATIQAIGVAQQAANVAATLKG), Cec4-8 (GWVKKIGKKIERVGQNTRDATIQVIGVAQQAANVAATLKG), and FITC-Cec4 (FITC-GWLKKIGKKIERVGQNTRDATIQAIGVAQQAANVAATLKG) were synthesized by solid-phase chemical synthesis by Gil Biochemical Co., Ltd., Shanghai. The purity (HPLC) was >95%. They were dissolved to 10 mg/mL with Distillation-Distillation H2O (ddH2O) and stored at −80 °C for further analysis.
Detecting the minimum inhibitory concentration (MIC) value and circular dichroism spectroscopy (CD) analysis
Evaluation of the MIC of synthetic peptides was performed using broth microdilution techniques according to the reference.24 The peptide Cec4 was dissolved and their secondary structure were detected according to the reference.25 The noise reduction processing was performed on all the maps after scanning with the software J7STDANL.
Detection of leakage of 260 nm absorbing material
The reference method was slightly modified,26 taking the logarithmic growth phase of A. baumannii (ATCC19606) as detected bacteria. 5 mL bacterial suspension (1×106 CFU/mL) was treated with different concentrations of antimicrobial peptides (1/4× MIC, 1/2× MIC, 1× MIC, 3× MIC). After being placed at 37 °C and 150 rmp, aliquots (0.5 mL) of the treated bacterial suspension were removed at different intervals (0 h, 0.5 h, 1 h, 2 h, 4 h, 6 h, 8 h). The aliquots were centrifuged and the absorbance of supernatant was measured at 260 nm, using phosphate buffer saline (PBS) as the control.
Transmission electron microscope
The logarithmic growth of A. baumannii (ATCC19606), diluted to 1.0×106 CFU/mL with MHB medium and treated with 1× MIC Cec4, was measured using PBS as the control. After being placed at 37 °C and 150 rmp, aliquots (0.2 mL) of the treated bacterial suspension were removed at different intervals (0 h, 1 h, and 3 h). The remaining steps were treated according to the reference.27
Fluorescence and confocal laser scanning microscopy (CLSM)
According to the method of reference,27 the logarithmic growth phase of A. baumannii (ATCC19606) as detected bacteria. 1× MIC FITC-Cec4 was added to the bacterial suspension, and 200 μL of the treated bacterial suspension was removed at regular intervals (30 min, 60 min, 90 min, and 120 min). At last, a volume of 20 μL of bacterial fixed with 4% paraformaldehyde at 4 °C overnight was observed by a fluorescence microscope or laser scanning confocal microscope.
Flow cytometry analysis
According to the method of,28 the A. baumannii in the logarithmic growth phase was taken. A volume of 200 μL of the prepared inoculum was added to 1× MIC of Cec4 peptide and then incubated at 37 °C for 60 min. The remaining steps were treated according to the reference.
DNA binding assay
Electrophoretic mobility shift was used to analyze the DNA binding ability of Cec4, as described in the reference.29
Biofilm formation and susceptibility assays
(1). Evaluation of biofilm formation ability of strains was performed by crystal violet staining as described in the literature.30 Briefly, strains were cultured in nutrient agar for 18–20 h and adjusted to 0.5 McFarland units (~1.5×108 CFU/mL) using Mueller-Hinton broth (MHB) medium. A 10 μL aliquot of each suspension was then diluted 1:20 in 190 μL of fresh MHB medium in 96-well polyvinylchloride microtiter plates and incubated at 37 °C for 24 h. The culture medium was subsequently removed, and wells were carefully washed with PBS three times to remove planktonic bacteria before refilling the wells with fresh MHB medium. Peptides and antibiotics were added at different concentrations, and plates were incubated at 37 °C for 24 h. After the removal of medium at the end of incubation, wells were rinsed by submerging the entire plate in a tub containing tap water. Biofilms were stained with 0.1% (wt/vol) crystal violet for 30 min. After staining, the dye was removed and the wells were washed with distilled water. The plates were dried for at least 1 h prior to addition of ethanol (95%) to solubilize the dye bound to the biofilm. The OD of biofilms was measured at 595 nm absorbance by using a microplate reader. The percentage of inhibition was calculated using the equation (1 – A595 of the test/A595 of non-treated control) ×100. The minimum biofilm inhibition concentration (MBIC90) was defined as the lowest agent concentration that showed 90% or more inhibition of the formation of biofilm. ATCC19606 was used as a reference strain, while un-inoculated LB medium was used as a negative control.31 All experiments were carried out in triplicate.
(2). The minimum biofilm eradication concentrations (MBEC) of Cec4 for A. baumannii isolates were assessed with a micro dilution assay.32 Test strains were cultured in 96-well flat-bottomed cell culture plates for 24 hrs at 37 °C to form biofilms, and then treated with 64–2048 μg/mL Cec4 for 24 h; follow on incubation with fresh MHB medium for 24 hrs at 37 °C to restore residual bacteria. All assays were repeated in triplicate.
Hemolysis assay
Human blood was provided by healthy volunteers, and the use of healthy human blood was approved by the IRB of Guizhou Medical University. In addition, the donors provided written informed consent, in compliance with the Declaration of Helsinki. Healthy human blood was centrifuged at 2000 rpm for 5 mins, and then human red blood cells (RBC) were washed three times with PBS. An 8% (v/v) RBC suspension was prepared in PBS, and 50 μL of the solution was transferred to a 96-well plate. Then, 50 μL of different concentrations of Cec4 solution were added. PBS and Triton X-100 (0.1%) were used as a negative and a positive control, respectively. Finally, the hemolysis of the peptide Cec4 was evaluated by measuring the absorbance at OD405 with a microplate reader. Experiments were done in triplicate as described in the reference.29
Statistical analyses
Comparison of data between groups using two-tailed Student’s t-test (*P<0.05, **P<0.01).
Results
Circular dichroism (CD) spectroscopy analysis
In order to study the relationship between the structure and function of Cec4, two mutants of the cecropin family member in Musca domestica (Cec4-7 and Cec4-8) with only an amino acid difference with Cec4 were synthesized (Figure 1A). The secondary structures of synthesized peptides in aqueous (PBS) and microbial membrane simulation solvent (30 mM Sodium dodecyl sulfate, SDS) were investigated with CD spectroscopy. Results showed that the peptides Cec4, Cec4-7 and Cec4-8 demonstrated an antiparallel β-fold conformation in the aqueous solvent, and the contents were 97.20%, 93.3% and 84.4% (Figure 1B). It was found that Cec4, Cec4-7 and Cec4-8 have an increase in α-helix content in the presence of microbial membrane simulation solvent SDS from 41.6%, 18.0% and 39.0% up to 99.7%, 99.2% and 99.7% respectively (Figure 1C). The peptides changed their conformation in the presence of microbial membrane simulation solvent (Table S1). It has been demonstrated that the MIC of Cec4 against A. baumannii was 4 µg/mL, and the MIC values of Cec4-7 and Cec4-8 are more than 256 µg/mL (Table 1). Therefore, the Cec4 more easily forms α-helix structures in both aqueous and hydrophobic environments, which was consistent with the MIC analysis.
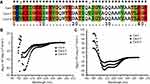 |
Figure 1 Sequence alignment and circular dichroism (CD) spectra of peptides. (A) Multi-sequence alignment of peptides performed using ClustalW (http://embnet.vital-it.ch/software/ClustalW.html). CD spectra of Cec4, Cec4-7, and Cec4-8 in PBS (B) and the microbial membrane-mimicking environment (25 mM SDS micelles solution) (C). |
 |
Table 1 Determination of MIC values of peptides against selected strains (μg/mL) |
Bacterial membrane disruption activity of Cec4
It has been confirmed that the potent antibacterial activity of the Cec4 is not just against standard A. baumannii (ATCC19606), but also against MRAB and PRAB, and the MICs were 4 μg/mL (Table 1). In order to gain insight into the antibacterial mechanism of Cec4, a variety of experiments were carried out. Firstly, damage to A. baumannii outer membrane by Cec4 was evaluated by measuring 260 nm absorbing material of A. baumannii mixed with Cec4. As demonstrated in Figure 2, dose-dependent increase is very significant in the maximum absorbance at 260 nm of bacteria mixed with Cec4. The leakage rates of ¼ MIC Cec4 and 3× MIC Cec4 groups were concentration-dependent (P˂0.05), which indicated that the destruction of peptide Cec4 on the bacterial membrane was in dose and time dependent manner. These results indicate that Cec4 may target the A. baumannii outer membrane, causing intracellular material to leak to the outer membrane and increase the maximum absorbance at 260 nm.
 |
Figure 2 Leakage of 260 nm-absorbing materials from cell suspensions of A.baumannii exposed to Cec4 (*P<0.05, **P<0.01). |
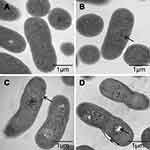
To explore the positional relationship of Cec4 and A. baumannii, the fluorescence microscope found that more and more bacteria were tagged with FITC-Cec4 as time increased (Figure S1). At this point, however, it was not clear whether the antimicrobial peptides had entered the bacterial cell. To further confirm the damage of Cec4 to A. baumannii membranes, A. baumannii (19606) treated with Cec4 (1× MIC) was observed by transmission electron microscopy (TEM). TEM micrographs show that the outer cell membrane and inner cell membrane of untreated bacteria are intact and the cell walls are clearly visible. More importantly, the electron density in the cytoplasmic region is very homogeneous. (Figure 3A). However, significant damage to bacterial cells was observed after exposure to Cec4. After treatment with a low concentration of Cec4 (1× MIC) for 0.5 hrs, a high electron density condensed substance was observed in the cells (Figure 3B). After A. baumannii were treated with Cec4 (1× MIC) for 1 h, the bacteria exhibited a foamy disintegrating membrane and an enlarged periplasmic space, and the surface of the bacteria had filament fragments (Figure 3C). After treatment with Cec4 (1× MIC) for 3 hrs, complete loss of integrity of cell membrane and cell wall was clearly observed. Due to leakage of cytoplasmic contents, heterogeneous cytoplasmic density and dissolved cell membrane structure were also observed (Figure 3C and D). These data confirm the destruction of A. baumannii membrane by Cec4.
It is known that antimicrobial peptide Cec4 can destroy the cell membrane; whether the Cec4 will get into the bacterial cells and accumulate intracellular is still elusive. Therefore, fluorescence microscopy and LSCM-related experiments were used (Figure S1). As shown in Figure 4A–C, a small fraction of the bacteria was labeled and it entered the interior of the bacteria in 30 min; when applied for 60 min (Figure 4D–F), the amount of peptide Cec4 entering the bacterial cells increased. This suggests that the peptide Cec4 also has a binding specificity with the bacterial membrane.
DNA-binding property and flow cytometry analysis
It is reported that the antibacterial effect of AMP is not restricted to membrane perforation, so the possibility of other intracellular targets (such as DNA) is explored by detecting the DNA binding properties of the peptide Cec4. As shown in Figure S2, the peptide Cec4 was incapable of binding to plasmid DNA, even though it was at a high concentration (64× MIC) and treated with the plasmid DNA for 3 h.
Flow cytometry was used to assess the effect of peptide Cec4 on bacterial cell cycle. In the control group, the number of A. baumannii cells in G1 phase accounted for 33.47%, and S phase accounted for 30.47% (Figure 5A). After peptide Cec4 was applied for 30 min, the number of A. baumannii cells in G1 accounted for 53.36%, which was higher than that of the control. The number of A. baumannii cells in S phase then accounted for 33.26%, which was increased by 9.15% relative to the control (Figure 5B). After the addition of the peptide Cec4, the number of A. baumannii cells in the G1 phase was significantly increased, indicating that most of the cells stopped in the G1 phase and no longer continued to the R phase.
 |
Figure 5 Effects of antimicrobial peptide Cec4 on the cell cycle of A. baumannii (A) Untreated A. baumannii cells; (B) 0.5 h after peptide Cec4-treated A. baumannii. |
The quantification of Cec4 against biofilms; Cec4 has no cell toxicity
A. baumannii easily forms biofilm, and weakening the biofilm formation will help to eliminate this kind of drug-resistant bacteria. To investigate the scavenging effect of Cec4 on established biofilms, the amount of biofilms of standard and clinically isolated A. baumannii was determined by crystal violet staining.As observed in Figure 6, results show that the peptide Cec4 at 0.5 μg/mL can inhibit the biofilm formation of A. baumannii. Remarkably, with the concentrations of Cec4 at 4 μg/mL, 2 μg/mL and 4 μg/mL, they inhibited 50% of the biofilm formation of standard A. baumannii, MRAB and PRAB respectively. Peptide Cec4 significantly reduced standard A. baumannii and MRAB biofilms compared to the results with PRAB.
For the sake of eliminating drug-resistant bacteria more completely, biofilm inhibitor must not only reduce the formation of biofilms that inhibit resistant bacteria in the initial stage, but they must also eradicate the mature biofilms that have already been formed. The capability of Cec4 to eliminate established biofilm of A. baumannii was examined. As shown in Figure 6, Cec4 disrupted mature (24 hr) biofilms of standard A. baumannii and MRAB more significantly than it did PRAB. When treated with Cec4 (64 μg/mL), the biofilms formed by standard A. baumannii and MRAB could not grow again. Meanwhile, Cec4 (128 μg/mL) can restrain the regrowth of biofilm formed by PRAB (Table 2). It was demonstrated that Cec4 has a concentration-dependent biofilm-disrupting activity against A. baumannii. Further biofilm eradication experiments found that Cec4 (128 μg/mL) can eradicate the biofilm formation of standard A. baumannii and MRAB, and Cec4 (256 μg/mL) can eradicate the biofilm of PRAB (p<0.05) (Table 2). Collectively, the data demonstrate that peptide Cec4 is effective in penetrating and disrupting adherent standard A. baumannii and MRAB biofilms more effectively than for PRAB in concentration-dependent manner.
 |
Table 2 Determination of MBIC and MBRC values of Cec4 against selected strains (μg/mL) |
The cytotoxicity of drugs is an important parameter to be evaluated in the development of antibiotics, especially drugs whose target is cell membrane. As shown in Figure 7, results indicated very low hemolysis (less than 4%) with Cec4 even in high concentration (600 μg/mL). However, 0.1% Triton X-100 can completely lyse human red blood cells (100% hemolysis).
Discussion
The antibiotic resistance of A. baumannii is becoming more and more serious. Polymyxin B and colistin, which target the cell membrane, are considered to be the last line of defense against drug-resistant bacteria, but these drugs have large side effects and drug resistance to such drugs is gradually increasing. In response to the threat of A. baumannii, some research groups have tried to kill multi-drug resistant A. baumannii using AMP.34 Although AMPs were discovered several decades ago, the mechanism by which they kill bacteria is still controversial. It has been several decades since the first discovered AMPs, but the exact mechanism of them killing bacteria is still elusive. It has been reported that the antibacterial activity of antimicrobial peptides is closely related to its amphiphilic structure. The amphiphilic conformation is reflected as the α-helix or β-sheet, and it is interesting to note that a conformational change occurs only when the antimicrobial peptide is in the membrane environment.35 When such a change occurs, the hydrophilic amino acid residue and the hydrophobic amino acid residue of the antimicrobial peptide are no longer closely linked, so that the antimicrobial peptide can function better with the cell membrane and cause opening of ion channels or pores on the membrane. In this study, Cec4-8 was obtained by replacing the hydrophobic leucine at position 3 of Cec4 with hydrophobic valine, resulting in an increase in the inhibitory concentration from the original 4 μg/mL to more than 256 μg/mL, indicating that leucine is a key amino acid in antimicrobial peptide activity. The secondary structure content of Cec4 and Cec4-8 in different buffers was detected by circular dichroism. The results showed that Cec4 and Cec4-8 had little difference in their secondary structure in different environments. The α-helix contents of Cec4 and Cec4-8 in a hydrophobic environment both are 99.7%, but there is a big difference in antibacterial activity. This suggests that the high content of α-helix may not be seen as the only factor for antibacterial activity, which provides a theoretical basis for the subsequent modification and design of antimicrobial peptides.
The ability of the new peptide to penetrate the A. baumannii membrane was assessed by detecting the accumulation of peptides into the A. baumannii cell membrane and causing the release of intracellular components of A. baumannii (Figure 1 and Table S1).26 These findings indicate that Cec4 targets the bacterial outer membrane, causing intracellular material to leak into the outer membrane. Over time, the maximum absorbance value at A260nm is proportional to the damage of the cell membrane structure. Membrane permeation is not the only destructive factor that governs the activity of AMPs against microbial cells. Several reports indicated that AMPs could have multiple inhibitory effects by affecting the functions of other cellular constituents, such as the cell wall, DNA, RNA, and cellular proteins.36 However, membrane permeation is not the only way of AMP killing microorganisms. Some reports indicate that AMPs can have multiple inhibitory effects by affecting the function of other cellular components (such as cell walls, DNA, RNA, and cellular proteins). In addition to membrane permeability, the binding bacterial DNA ability of Cec4 peptide was evaluated. The results showed that Cec4 hardly bound to bacterial genomic DNA (Figure S2). The data indicates that DNA is not one of the targets that cause the antibacterial activity of Cec4. This phenomenon is consistent with the cationic antibacterial peptide magainin of amphibious skin. Even at concentration up to 80 M, it does not have the ability to bind DNA.29 In order to analyze other effects of antimicrobial peptide Cec4, this study used flow cytometry to detect the bacterial cell cycle of Cec4 against A. baumannii. A. baumannii is a prokaryote, and the bacterial cell cycle is divided into three phases (I, R, and D). Phase I is the preparation period for cell division to DNA replication, corresponding to the G1 phase of the eukaryotic cell cycle. In the R phase DNA is replicated, and this corresponds to the S phase of the eukaryotic cell cycle.17 After the addition of the peptide Cec4, the results showed that the Cec4 blocked more bacteria in the G1 phase, and the bacteria entering the G2/M phase were significantly reduced.
The formation of bacterial biofilm is considered to be one of the current challenges for drug-resistant bacteria, and biofilms play a crucial role in the pathogenesis of A. baumannii infection. Common antibiotics mainly inhibit the metabolism and macromolecular synthesis of proliferating bacteria. In fact, bacteria in biofilms either grow slowly or are in dormancy, leading to stronger drug resistance.8 In fact, it was found that gentamicin and tobramycin can induce the formation of P. aeruginosa PAO1 biofilm at sub-inhibitory concentrations. Compared with the control, the biofilm can increase by 100–240%.33 However, our data show that Cec4 can not only remove free A. baumannii at lower concentrations, but also inhibit the formation of A. baumannii biofilm and remove the biofilm formed A. baumannii.
AMP has a good effect on some clinical super bacteria such as P. aeruginosa and A. baumannii, but their clinical research is not smooth due to their poor stability in vivo and possible toxic effects such as hemolysis or immunomodulation. However, the Cec4 we studied, even at high concentrations (300 μg/mL), still had very low hemolytic reaction (less than 4%).
Conclusion
In conclusion, the antibacterial mechanism of peptide Cec4 against A. baumannii has two main aspects. Our results showed that peptide Cec4 not only acts on the cell membrane but also intracellular, suggesting that the antimicrobial peptide may have a multi-target mechanism, leading to rapid death of bacteria. Therefore, the risk of antimicrobial resistance can be reduced, providing an option for the exploitation of new clinical drugs, and also providing a theoretical basis for the research on new antimicrobial peptide mechanisms.
Acknowledgments
This research is funded by the National Natural Science Foundation of China (No. 81660347), the Guizhou Provincial Science and Technology Plan Project ([2017]1154), Youth Science and Technology Talents Growth Project of Guizhou Provincial Department of Education ([2016]148), Key Technologies R&D Program for Science and Technology Department of Guizhou Province ([2019]2823), and Doctoral Fund of Guizhou Medical University ([2015]002). Funders had no role in study design, data collection or analysis, preparation of the manuscript or the decision to publish it. We also want to thank Emma Taylor for proofreading the paper.
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Heidary M, Salimi Chirani A, Khoshnood S, et al. Molecular detection of aminoglycoside-modifying enzyme genes in Acinetobacter baumannii clinical isolates. Acta Microbiol Immunol Hung. 2017;64(2):143–150. doi:10.1556/030.63.2016.022
2. Kroger C, Kary SC, Schauer K, Cameron AD. Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes (Basel). 2016;8(1):12. doi:10.3390/genes8010012
3. Sader HS, Farrell DJ, Flamm RK, Jones RN. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalised with pneumonia in US and European hospitals: results from the SENTRY Antimicrobial Surveillance Program, 2009–2012. Int J Antimicrob Agents. 2014;43(4):328–334. doi:10.1016/j.ijantimicag.2014.01.007
4. Asadollahi K, Alizadeh E, Akbari M, et al. The role of bla(OXA-like carbapenemase) and their insertion sequences (ISS) in the induction of resistance against carbapenem antibiotics among Acinetobacter baumannii isolates in Tehran hospitals. Roum Arch Microbiol Immunol. 2011;70(4):153–158.
5. Bonnin RA, Nordmann P, Poirel L. Screening and deciphering antibiotic resistance in Acinetobacter baumannii: a state of the art. Expert Rev Anti Infect Ther. 2013;11(6):571–583. doi:10.1586/eri.13.38
6. Boll JM, Crofts AA, Peters K, et al. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc Natl Acad Sci U S A. 2016;113(41):E6228–E6237. doi:10.1073/pnas.1611594113
7. Poirel L, Bonnin RA, Nordmann P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life. 2011;63(12):1061–1067. doi:10.1002/iub.532
8. Gayoso CM, Mateos J, Mendez JA, et al. Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J Proteome Res. 2014;13(2):460–476. doi:10.1021/pr400603f
9. China Antimicrobial Resistance Surveillance S. [Surveillance of bacterial resistance in children and newborns across China from 2014 to 2017]. Zhonghua Yi Xue Za Zhi. 2018;98(40):3279–3287. doi:10.3760/cma.j.issn.0376-2491.2018.40.013
10. Mangoni ML, Bhunia A. Editorial: antimicrobial peptides in medicinal chemistry: advances and applications. Curr Top Med Chem. 2016;16(1):2–3.
11. Lazar V, Martins A, Spohn R, et al. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat Microbiol. 2018;3(6):718–731. doi:10.1038/s41564-018-0164-0
12. Pane K, Durante L, Crescenzi O, et al. Antimicrobial potency of cationic antimicrobial peptides can be predicted from their amino acid composition: application to the detection of “cryptic” antimicrobial peptides. J Theor Biol. 2017;419:254–265. doi:10.1016/j.jtbi.2017.02.012
13. Le CF, Fang CM, Sekaran SD. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob Agents Chemother. 2017;61(4). doi:10.1128/AAC.02340-16
14. Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10(3):310–315. doi:10.1038/nm996
15. Gui S, Li R, Feng Y, Wang S. Transmission electron microscopic morphological study and flow cytometric viability assessment of Acinetobacter baumannii susceptible to Musca domestica cecropin. Scientific World Journal. 2014;2014:657536. doi:10.1155/2014/657536
16. Lu X, Shen J, Jin X, et al. Bactericidal activity of Musca domestica cecropin (Mdc) on multidrug-resistant clinical isolate of Escherichia coli. Appl Microbiol Biotechnol. 2012;95(4):939–945. doi:10.1007/s00253-011-3793-2
17. Sang M, Wei H, Zhang J, et al. Expression and characterization of the antimicrobial peptide ABP-dHC-cecropin A in the methylotrophic yeast Pichia pastoris. Protein Expr Purif. 2017;140:44–51. doi:10.1016/j.pep.2017.08.001
18. Thandar M, Lood R, Winer BY, Deutsch DR, Euler CW, Fischetti VA. Novel engineered peptides of a phage lysin as effective antimicrobials against multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2016;60(5):2671–2679. doi:10.1128/AAC.02972-15
19. Alarcon T, Lopez-Hernandez S, Andreu D, Saugar JM, Rivas L, Lopez-Brea M. In vitro activity of CA(1-8)M(1-18), a synthetic cecropin A-melittin hybrid peptide, against multiresistant Acinetobacter baumannii strains. Rev Esp Quimioter. 2001;14(2):184–190.
20. Long H, Yang J, Peng J, Jianwei W. Antimicrobial activity of Musca domestica cecropin - 4 (mdCec 4) against Acinetobacter baumannii. Chin J Microbiol Immunol. 2017;37(12):891–896. doi:10.3760/cma.j.issn.0254-5101.2017.12.002
21. Liang Y, Wang JX, Zhao XF, Du XJ, Xue JF. Molecular cloning and characterization of cecropin from the housefly (Musca domestica), and its expression in Escherichia coli. Dev Comp Immunol. 2006;30(3):249–257. doi:10.1016/j.dci.2005.04.005
22. Jin X, Mei H, Li X, et al. Apoptosis-inducing activity of the antimicrobial peptide cecropin of Musca domestica in human hepatocellular carcinoma cell line BEL-7402 and the possible mechanism. Acta Biochim Biophys Sin. 2010;42(4):259–265. doi:10.1093/abbs/gmq021
23. Lee HW, Koh YM, Kim J, et al. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin Microbiol Infect. 2008;14(1):49–54. doi:10.1111/j.1469-0691.2007.01842.x
24. Sahu C, Jain V, Mishra P, Prasad KN. Clinical and laboratory standards institute versus European committee for antimicrobial susceptibility testing guidelines for interpretation of carbapenem antimicrobial susceptibility results for Escherichia coli in urinary tract infection (UTI). J Lab Physicians. 2018;10(3):289–293. doi:10.4103/JLP.JLP_176_17
25. Ong ZY, Cheng J, Huang Y, et al. Effect of stereochemistry, chain length and sequence pattern on antimicrobial properties of short synthetic beta-sheet forming peptide amphiphiles. Biomaterials. 2014;35(4):1315–1325. doi:10.1016/j.biomaterials.2013.10.053
26. Patra JK, Baek KH. Antibacterial activity and action mechanism of the Essential Oil from Enteromorpha linza L. against foodborne pathogenic bacteria. Molecules. 2016;21(3):388. doi:10.3390/molecules21030388
27. Schneider VA, Coorens M, Ordonez SR, et al. Imaging the antimicrobial mechanism(s) of cathelicidin-2. Sci Rep. 2016;6:32948. doi:10.1038/srep32948
28. Shen Y, Vignali P, Wang R. Rapid profiling cell cycle by flow cytometry using concurrent staining of DNA and mitotic markers. Bio Protoc. 2017;7(16). doi:10.21769/BioProtoc.2517
29. Mohamed MF, Hamed MI, Panitch A, Seleem MN. Targeting methicillin-resistant Staphylococcus aureus with short salt-resistant synthetic peptides. Antimicrob Agents Chemother. 2014;58(7):4113–4122. doi:10.1128/AAC.02578-14
30. Wei GX, Campagna AN, Bobek LA. Effect of MUC7 peptides on the growth of bacteria and on Streptococcus mutans biofilm. J Antimicrob Chemother. 2006;57(6):1100–1109. doi:10.1093/jac/dkl120
31. Kong L, Qi X, Huang S, Chen S, Wu Y, Zhao L. Theaflavins inhibit pathogenic properties of P. gingivalis and MMPs production in P. gingivalis-stimulated human gingival fibroblasts. Arch Oral Biol. 2015;60(1):12–22. doi:10.1016/j.archoralbio.2014.08.019
32. Ceri H, Olson M, Morck D, et al. The MBEC Assay System: multiple equivalent biofilms for antibiotic and biocide susceptibility testing. Methods Enzymol. 2001;337:377–385.
33. Mohamed MF, Brezden A, Mohammad H, Chmielewski J, Seleem MN. A short D-enantiomeric antimicrobial peptide with potent immunomodulatory and antibiofilm activity against multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Sci Rep. 2017;7(1):6953. doi:10.1038/s41598-017-07440-0
34. Tucker AT, Leonard SP, DuBois CD, et al. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell. 2018;172(3):618–628 e613. doi:10.1016/j.cell.2017.12.009
35. Dennison SR, Harris F, Mura M, Morton LH, Zvelindovsky A, Phoenix DA. A novel form of bacterial resistance to the action of eukaryotic host defense peptides, the use of a lipid receptor. Biochemistry. 2013;52(35):6021–6029. doi:10.1021/bi400719j
36. Fjell CD, Hiss JA, Hancock RE, Schneider G. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov. 2012;11(1):37–51. doi:10.1038/nrd3591
Supplementary materials
 |
Table S1 Secondary structure prediction of AMPs (190–260 nm). |
 |
Figure S1 Fluorescence microscopy was used to observe the location of antimicrobial peptide and bacteria. (A–D) 1× MIC Cec4 treated A. baumannii 30 min, 60 min, 90 min, 120 min respectively. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.