Back to Journals » OncoTargets and Therapy » Volume 12
Androgen upregulates the palmitoylation of eIF3L in human prostate LNCaP cells
Authors Cui L, Liu M, Lai S, Hou H, Diao T, Zhang D, Wang M, Zhang Y, Wang J
Received 18 November 2018
Accepted for publication 15 April 2019
Published 5 June 2019 Volume 2019:12 Pages 4451—4459
DOI https://doi.org/10.2147/OTT.S193480
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Luwei Cui,1,2 Ming Liu,2 Shicong Lai,2,3 Huimin Hou,2,3 Tongxiang Diao,1,2 Dalei Zhang,2 Miao Wang,2,3 Yaoguang Zhang,1,2 Jianye Wang1,2
1Peking University Fifth School of Clinical Medicine, Beijing, People’s Republic of China; 2Department of Urology, Beijing Hospital, National Center of Gerontology, Beijing, People’s Republic of China; 3Graduate School of Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, People’s Republic of China
Background: Prostate cancer is the second leading cause of cancer-related deaths in Western countries. Most patients diagnosed with advanced prostate cancer can be treated with the main treatment: androgen deprivation therapy (ADT). The androgen receptor (AR) signaling axis plays a pivotal role in the progression of prostate cancer. However, most patients can ultimately progress to the castration-resistant prostate cancer (CRPC) stage within 2 years. At this stage, drugs targeting the AR signaling axis, including enzalutamide and abiraterone acetate, cannot prevent the progression of prostate cancer, thus predicting a poor prognosis. The molecular mechanism lies in the aberrant AR reactivation, which exhibits an adaptive response to ADT, such as the presence of AR splice variants. Thus, CRPC treatment remains a challenge.
Purpose: In addition to the AR axis, a mechanism leading to this progression should be determined. The present study mainly compared palmitoylated proteins between androgen-treated LNCaP cells and non-treated LNCaP cells by palmitoylome profiling, to illustrate the changes at proteomic levels.
Materials and methods: To screen the androgen-induced palmitoylated proteins, we conducted proteomic experiments using clickable palmitate probe (Alk-C16) between three individual pairs of androgen-treated and non-treated LNCaP cells.
Results: We identified 4351 unique peptides corresponding to 835 proteins, among them a number of these identified proteins were palmitoylated proteins, particularly eIF3L. Androgen treatment significantly increased the palmitoylation level of eIF3L, an individual subunit of eIF3. As an initiation factor, eIF3L plays a pivotal role in the translation of mRNAs encoding growth-promoting proteins by enhancing translation rates, thus controlling cell proliferation.
Conclusion: In this study, we demonstrated that the regulation of eIF3L palmitoylation may provide new directions for the therapy of prostate cancer. Moreover, the increased level of androgen-induced eIF3L may be used as a biomarker for the diagnosis of early-stage prostate cancer.
Keywords: androgen, palmitoylation, eIF3L, prostate cancer, biomarker
Introduction
Updated statistics in 2018 indicate that prostate cancer remains the most commonly diagnosed malignant tumors in men. A total of 164,690 new cases of prostate cancer have been diagnosed and 29,430 related-deaths have been reported in the United States.1 An estimated 450,000 males have been diagnosed with prostate cancer in Europe, comprising a large proportion of the overall burden of cancers worldwide.2 Prostate cancer is the most common noncutaneous malignancy diagnosed in American men and androgen-dependent malignant tumor among males.3,4 The interaction between androgen and androgen receptor (AR) is the cornerstone that leads to progression, thus drugs targeting the AR signaling axis remain the principal therapy for advanced prostate cancer.5 Androgen deprivation therapy (ADT) is initially effective, leading to a marked decrease in PSA. However, the majority of the patients with advanced prostate cancer can progress and become refractory to ADT owing to aberrant AR activation, which is castration-resistant prostate cancer. The median survival at this stage is historically less than 2 years.6,7 The associated mechanisms include the occurrence of AR splice variants,8 inducing 20–40% of the patients to respond poorly to enzalutamide or abiraterone, both of which are large breakthroughs in the treatment of metastatic castration-resistant prostate cancer (CRPC). Secondary resistance to these drugs is regarded as an adaptive change in prostate cancer cells. Thus, therapeutic methods targeting the novel mechanism need to be developed to meet the clinical need of patients with metastatic CRPC.
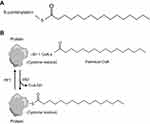
Posttranslational modification helps provide protein with additional functions, among which protein fatty acylation is rather common.9 Protein fatty acylation is the covalent attachment of lipids onto proteins, controlling protein–protein and protein–membrane interactions, which play a pivotal role in modulating the biological functions of proteins and mediating their targeting for activation.10,11 Thus, this modification is essential for cells to maintain homeostasis and to respond instantly to extracellular signals! Several forms of protein fatty acylation exist in eukaryocytes, which primarily include N-myristoylation and S-palmitoylation. S-palmitoylation is the addition of a 16-carbon saturated fatty acid, the so-called palmitate, to cysteine residues via a labile thioester linkage12 (Figure 1A). Among these modifications, palmitoylation is a common form of protein fatty acylation, by which protein hydrophobicity is efficiently increased to facilitate membrane association. In addition, palmitoylation is closely associated with the regulation of protein trafficking, stability and activity.13,14 Several studies have demonstrated that protein palmitoylation is associated with the development of disease, tumorigenesis and prognosis. Studies have proved the presence of aberrant palmitoylation in Huntington disease, caused by a CAG expansion in the huntingtin (HTT) gene. The palmitoyl acyltransferases for HTT are disturbed, resulting in reduced palmitoylation of HTT and exhibiting itself with motor, cognitive and psychiatric deficits.15 Moreover, Ras tumorigenesis is also regulated by palmitoylation. The Ras family of proteins widely consists of cancer drivers and plays a role in signaling, cell proliferation, differentiation and survival.16,17 Two reports in 2005 indicated that the presence of Ras was controlled by a transport cycle via palmitoylation and depalmitoylation,18,19 thus determining its location to membrane and subsequent oncogenic signaling (Figure 1B). Therefore, targeting and disrupting membrane interactions of specific Ras isoforms represent a significant therapeutic strategy for regulating Ras-driven tumorigenesis. Nature in 2017 indicated that palmitoylation-mediated activation of MC1R could prevent melanomagenesis.20 From the aforementioned discussion, palmitoylation plays a key role in membrane location, subsequent signaling pathway and tumorigenesis. Considering the limitations of the current drug therapy for CRPC, we explore palmitoylated proteins in androgen-treated LNCaP cells and non-treated LNCaP cells, to reveal the mechanism of prostate cancer progression by protein palmitoylation and identify a new drug target!
Click-chemistry-based chemical probes for the detection of protein palmitoylation hasten the discovery of novel palmitoylated proteins and elucidate their biological functions.21 The combination of mass-spectrometry-based quantitative proteomics with click chemistry probes can be used to explore the dynamics of protein palmitoylation under different physiological or pathological conditions.22,23 In the present study, we used the chemical tool and compared the palmitoylated proteins between androgen-treated LNCaP cells and non-treated LNCaP cells in the palmitoylome profiles. We found that the palmitoylation level of eIF3L was high in both androgen-treated and non-treated LNCaP cells, and androgen treatment significantly increased the palmitoylation level of eIF3L. Considering that eIF3L serves as an initiation factor and palmitoylated eIF3L might cooperate with the initiation complex and enhance the translation of mRNAs,24 we demonstrated that androgen-induced palmitoylation of eIF3L might play a pivotal role in AR gene expression and cancer progression, making it possible to provide new directions for therapeutic targets. Moreover, the high level of palmitoylated eIF3L induced by androgen may serve as a novel biomarker in the diagnosis of early prostate cancer.
Materials and methods
S-palmitoylation assay
Prior to the experiments, LNCaP cells were seeded with complete media onto 6 cm-dishes (5×105 cells/dish) and incubated for 48 hrs. We purchased LNCaP cells from American Type Culture Collection, which were grown in RPMI-1640 media supplemented with 10% FBS and incubated in a 5% CO2 humidified chamber at 37°C for 48 hrs before any experiment. The cells were then treated with R1881 (10 nM) or DMSO in RPMI-1640 media supplement with 0.5% FBS. After 24 hrs, the cells were harvested for S-palmitoylation assay.
The S-palmitoylation assay was performed in accordance with the protocols described previously25 and slightly modified. The following are the inclusive reagents needed in this experiment: Nethylmaleimide (NEM, Sigma), Protein (A/G) UltraLink Resin (Thermo), Rabbit anti-eIF3L (1:2000, Protein tech), hydroxylamine (Sigma), tris (2-carcboxy-ethyl) phosphine hydrochloride (TCEP, Sigma), tris [(1-benzyl-1H-1, 2, 3-triazol-4-yl) methyl] amine (TBTA, Sigma), Thiopropyl Sepharose 6B (Sigma) and azide-agarose beads (Nanocs). LNCaP cells were homogenized in lysis buffer (10 mM sodium phosphate, 2 mM Na2EDTA, 0.32 M sucrose, 1% Triton X-100, 50 mM NEM and Pierce protease and phosphatase inhibitor cocktail, pH 7.4) for 30 mins. The lysates were then immunoprecipitated using protein A/G resin preloaded with eIF3L antibody overnight. The following day, the protein A/G resin was washed three times and incubated with elution buffer (1% SDS, 10 mM sodium phosphate, 2 mM Na2EDTA, 0.32 M sucrose) at 50°C for 5 mins to release eIF3L proteins. Eluted samples were divided into two equal portions—one treated with 1 M hydroxylamine and the other with 1 M Tris·HCl (pH 7.4) (as a control) with the presence of activated thiol-sepharose 4B (Sigma). After incubation for 2 hrs at room temperature, the protein A/G resin was washed with a wash buffer (10 mM sodium phosphate, 2 mM Na2EDTA, 0.32 M sucrose, 1% Triton X-100, 500 mM NaCl and 0.2% SDS) three times. Western blot analyses were performed to determine the presence of eIF3L protein.
Metabolic labeling and click chemistry accumulation
Before the experiments, the LNCaP cells were seeded with complete media onto 6-cm dishes (5×105 cells/dish) and then incubated for 48 hrs. Metabolic labeling and click chemistry were performed in accordance with the protocols previously described26 and then modified slightly. The ω-alkynyl fatty acid analog, Alk-C16, was dissolved in DMSO to generate 50 mM stock solutions and stored at −80°C. Prior to cell treatment, Alk-C16 was dissolved in RPMI-1640 serum-free media supplement with 5% BSA (fatty acid free) at a final concentration of 100 μM. The fatty acid media solutions were sonicated for 15 mins at room temperature. The fatty acid media were then divided into two equal portions—one added with R1881 (10 nM) and the other added with DMSO as a control. The seeded LNCaP cells were washed with PBS once and added the two fatty acid media, respectively, for 24 hrs at 37°C/5% CO2. After incubation for 24 hrs, the cells were washed three times and homogenized in 500 μL lysis buffer (1% Nonidet P-40/150 mM NaCl/Pierce protease and phosphatase inhibitor cocktail/100 mM sodium phosphate, pH 7.5) for 30 mins at 4°C. The protein extracts were then subjected to the probe labeling reaction for 1 hr at room temperature, at final concentrations of the following reagents: 1 mM azide-agarose beads, 1 mM TCEP dissolved in water, 0.2 mM TBTA dissolved in DMSO/tert-butanol (20%/80%) and 1 mM CuSO4 in PBS. The order of addition of the reagents to the protein extracts is important for the reaction. With click chemistry, the Alk-C16-conjugated proteins were accumulated in the azide-agarose beads. The agarose beads were washed three times with the lysis buffer at room temperature. The proteins accumulated in the beads were subsequently digested for mass spectrometry.
nLC-MS/MS analysis
Proteomic profiling was performed on an EASY nLC1000 (Thermo Fisher) system with an LTQ-Orbitrap-Elite (Thermo Fisher) mass spectrometer. Tryptic peptides were desalted using 0.1% fluoroacetic acid and loaded on a trap column (PepMap C18, 300 μm×5 mm). The peptides were then eluted to a 3 μm fused silica capillary column (75 μm×500 mm). Finally, the peptides were separated at two gradients: the gradient from 100% mobile phase A (97.9% H2O, 2% acetonitrile, 0.1% formic acid) to 30% mobile phase B (80% acetonitrile, 19.02% H2O, 0.08% formic acid) in the first 80 mins and the mobile phase B from 30% to 100% in the next 10 mins. The eluted peptides were directly sprayed with a voltage of 1.6 kV into the on-line coupled LTQ-Orbitrap-Elite MS using a nano electro-spray ionization source equipped with a metal-coated nano-scale emitter. Mass spectra were obtained over a mass-to-charge ratio (m/z) range of 300–1800 Th at a resolving power of 30,000 at 400 m/z.
Database search
The recorded MS spectra were analyzed using MaxQuant Software (version 1.5.5.1). The MS/MS peak list file was performed by searching against a forward and reverse version of the UniProtKB/Swiss-Prot human database (generated from version 2017_05, human taxonomy, 20,316 entries). The cutoff of the false discovery rate for peptide and protein identification was set to 0.01, and only peptides with ≥7 amino acid residues were allowed for identification. Label-free quantitation (LFQ) was conducted using MaxQuant on identified razor and unique peptides to properly quantify identified proteins.27 Gene ontology (GO) analysis was performed using DAVID (NIH) configured to assign the identified proteins with cellular function GO terms.
Data processing and statistical analysis
Protein abundances normalized using the LFQ algorithm integrated in MaxQuant were Log2 transformed for further analyses. Filtering was conducted in Microsoft Excel 2010. DanteR (version 1.0.1.1) and Perseus (version 1.5.5.3) were used to perform different types of statistical analysis including Log2 transformation, correlation plot, statistical tests, imputation, P-value adjustment and volcano plot.27
Results
Design and synthesis of clickable palmitate probe (Alk-C16)
Both alkyne and azide analogs of palmitic acid were developed for click chemistry applications. In the current study, we chose the alkyne analog because it exhibited physicochemical properties similar to those of the wild type fatty acid carbon chain and maintained the hydrophobicity for membrane affinity.26 (Figure 2) We added the alkyne group at the ω-position of a fatty acid. The position is the terminal end farthest from the carboxyl functionality.26 Our ω-Alkynyl palmitate analog (Alk-C16) was synthesized from alcohols containing internal alkynes via a zipper reaction. In the reaction, an internal alkyne was isomerized to a terminal alkyne followed by Jones oxidation to yield the ω-alkynyl palmitate analog with high yield and purity.26
Proteomic experiments using clickable palmitate probe (Alk-C16)
To compare the palmitoylated proteins between androgen-treated LNCaP cells and non-treated LNCaP cells in the palmitoylome profiles, Alk-C16, a palmitate fatty acid analog, was metabolically incorporated onto cellular proteins and the proteins modified with Alk-C16 were analyzed using MS-based proteomics and an informatics-assisted label-free strategy. First, the LNCaP cells were added with Alk-C16 and then incubated for 24 hrs. When the cells were metabolically incorporated with Alk-C16, the cells were divided into two parts to check the androgen-induced palmitoylated proteins: one treated with androgen (R1881) and the other with DMSO as the control. After 24 hrs, the cells were harvested and lysated. Alk-C16 incorporated onto acylated proteins was chemoselectively ligated to azide agarose beads by a Cu1-catalyzed alkyne-azide [3+2] cycloaddition reaction (click chemistry). The conjugated proteins, which were theoretically palmitoylated proteins, were accumulated in azide agarose beads by ligation. The conjugated proteins were then digested on the agarose beads for MS and label-free quantitation.
Androgen promotion of eIF3L palmitoylation
We supplied three individual pairs of androgen-treated and non-treated LNCaP cells to screen the androgen-induced palmitoylated proteins. As a result, we identified 4351 unique peptides corresponding to 835 proteins (mascot score>2, P<0.05). Meanwhile, we filtered the outlier proteins and contaminant proteins. The identified proteins were then analyzed in LFQ by MaxQuant version. A number of these identified proteins were palmitoylated proteins, particularly the eIF3 initiation factors, including its subunits eIF3j, eIF3g, eIF3c, eIF3b, eIF3a and eIF3L. We have also compared the amino acid sequences of eIF3L of Homo sapiens (human) with the mass spectrum results of eIF3L, and they are consistent (Figures 3 and 4). The protein with the relatively higher LFQ activity was eIF3L, one subunit of eIF3, both in androgen-treated and in non-treated LNCaP cells. Among these 6 subunits of eIF3, the androgen-induced candidates were identified as their LFQ activities were significantly upregulated (fold changes>1.5, P<0.05) in triplicate samples of androgen-treated vs non-treated LNCaP cells. Notably, both eIF3a and eIF3L were identified as the candidates (Figure 5).
 | Figure 3 The mass spectrum results of eIF3L palmitoylation. |
 | Figure 4 The amino acid sequences of eIF3L of Homosapiens (human). The partial highlighted red sequences of eIF3L are consistent with the mass spectrum results by comparing Figure 3 with Figure 4. |
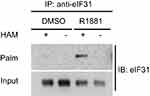
To strengthen the evidence, thiopropyl captivation of S-palmitoylated protein assay was further performed to explore whether androgen promotes the eIF3a and eIF3L palmitoylation levels in LNCaP cells, when the cells were treated with R1881 (androgen). The eIF3L palmitoylation level was significantly upregulated. This result was consistent with that of MS. We concluded from the aforementioned results that androgen upregulates the palmitoylation of eIF3L in human prostate LNCaP cells (Figure 6).
To further elucidate the regulatory effect of androgen on eIF3L palmitoylation, androgen (R1881) and abiraterone were added in the cultured LNCaP cells at the same time, but the concentration of abiraterone is varied from 0.1 µmol/L to 0.5 µmol/L; as the result shown in Figure 7, abiraterone could downregulate the eIF3L palmitoylation level by inhibiting androgen (R1881). From the above results, it can be seen that eIF3L palmitoylation is clearly regulated by androgen.
Discussion
Protein fatty acylation is critical in maintaining cellular homeostasis and regulating signaling pathway. We have shown that the clickable palmitate probe (Alk-C16) can be used for biochemical detection and cellular imaging of lipid-modified proteins. It facilitates our study of the dynamic behavior of lipid-modified proteins in various diseases and the determination of the pathological mechanism. It also has several advantages in cell culture for the detection of proteins that are palmitoylated under metabolic conditions such as drug treatment.22,23 Moreover, combined with quantitative proteomics, the clickable palmitate probe can quantify the palmitoylated proteins as well.
Protein synthesis is increasingly recognized as an important component in tumorigenesis and progression. Enhanced translation rates are closely associated with malignant transformation.28,29 Genome-wide analyses of mRNA translation by ribosome profiling indicates that translational control contributes significantly to overall gene expression.30 The initiation phase of protein synthesis is rate-limiting for most mRNAs. The translational components, particularly some initiation factors, play a pivotal role in the translational regulation.31
eIF3 is the largest of the initiation factors, comprising 13 non-identical protein subunits (eIF3a to m). It binds stably to 40S ribosomal subunits and promotes the binding of methionyl-tRNAi and mRNA to form the 40S initiation complex. The individual over-expression of 6 different eIF3 subunits is observed in various tumors, by enhancing the translation of mRNAs that encode growth-promoting proteins.32 The m7G cap-dependent scanning pathway of initiation involves the binding of a Met-tRNAi-40S ribosomal complex to the 5´-terminal region of an mRNA, followed by 5´ to 3´ scanning along the mRNA, recognition of the initiation codon, and junction of the 60S subunit.33–36 The 80S initiation complex formed can then enter the elongation phase of protein synthesis. The ribosomal complex, often called the 43S preinitiation complex or (43S PIC), comprises the 40S ribosome, eIF1,eIF1A, eIF3, and the ternary complex, eIF2-GTP-Met-tRNAi.
In our recent study, we detected the androgen-induced palmitoylated proteins by comparing the palmitoylome profile of androgen-treated LNCaP cells with that of non-treated cells. The results demonstrated that androgen significantly increased the level of palmitoylated eIF3L in androgen-treated LNCaP cells. This rapid response to extracellular signal: the androgen,is closely associated with the dynamic palmitoylated level of eIF3L. Prostate cancer is an androgen-driven solid tumor; thus, many therapeutic strategies target the AR signaling axis. However, when patients progress to the CRPC stage, the administration of drug therapy remains a challenge for urologists.
This preliminary study identified that androgen can induce the palmitoylation of eIF3L, which serves as the initiation factor and regulates protein expression and cell growth. Over-expression of eIF3 subunits is observed in many tumors by enhancing the translation of mRNAs. Further studies should be performed to confirm the correlation of palmitoylated eIF3L and cell proliferation. In addition, the correlation between the palmitoylated eIF3L and the prognosis of PCa patients was also worthy of further study. It is hoped that all the questions will be resolved with our proposed methods.
Conclusion
The results in this study indicate that other mechanisms may exist in the development and progression of prostate cancer. Androgen-induced protein palmitoylation may provide a new direction for the treatment of prostate cancer and the level of palmitoylated eIF3L may be a biomarker to detect early prostate cancer.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Grant 2016YFC0105505 & 2017YFC0910003 & 2017YFC0840100 & 2017YFC840102). The proteomic experiments were performed on the mass spectrometry platforms in State Key Laboratory of Membrane Biology, Institute of Zoology and Chinese Academy of Sciences. We thank Fei Xiao and Wen-qing Li, The Key Laboratory of Geriatrics, Beijing Hospital, National Center of Gerontology, for their guidance regarding this study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356–387. doi:10.1016/j.ejca.2018.07.005
3. Salinas CA, Tsodikov A, Ishak-Howard M, Cooney KA. Prostate cancer in young men: an important clinical entity. Nat Rev Urol. 2014;11(6):317–323. doi:10.1038/nrurol.2014.91
4. Huggins C, Hodges CV. Studies on prostatic cancer: I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. 1941. J Urol. 2002;168(1):9–12.
5. Kahn B, Collazo J, Kyprianou N. Androgen receptor as a driver of therapeutic resistance in advanced prostate cancer. Int J Biol Sci. 2014;10(6):588–595. doi:10.7150/ijbs.8671
6. Amaral TM, Macedo D, Fernandes I, Costa L. Castration-resistant prostate cancer: mechanisms, targets, and treatment. Prostate Cancer. 2012;2012:327253. doi:10.1155/2012/327253
7. Attar RM, Takimoto CH, Gottardis MM. Castration-resistant prostate cancer: locking up the molecular escape routes. Clin Cancer Res. 2009;15(10):3251–3255. doi:10.1158/1078-0432.CCR-08-1171
8. Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–1038. doi:10.1056/NEJMoa1315815
9. Iwanaga T, Tsutsumi R, Noritake J, Fukata Y, Fukata M. Dynamic protein palmitoylation in cellular signaling. Prog Lipid Res. 2009;48(3–4):117–127. doi:10.1016/j.plipres.2009.02.001
10. Greaves J, Chamberlain LH. Palmitoylation-dependent protein sorting. J Cell Biol. 2007;176(3):249–254. doi:10.1083/jcb.200610151
11. Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi:10.1146/annurev.bi.65.070196.001325
12. Aicart-Ramos C, Valero RA, Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta. 2011;1808(12):2981–2994. doi:10.1016/j.bbamem.2011.07.009
13. Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: regulation and function. Annu Rev Biochem. 2004;73:559–587. doi:10.1146/annurev.biochem.73.011303.073954
14. Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8(1):74–84. doi:10.1038/nrm2084
15. Sanders SS, Hayden MR. Aberrant palmitoylation in Huntington disease. Biochem Soc Trans. 2015;43(2):205–210. doi:10.1042/BST20140242
16. Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol. 2003;4(5):373–384. doi:10.1038/nrm1105
17. Castellano E, Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer. 2011;2(3):216–231. doi:10.1177/1947601911408081
18. Rocks O, Peyker A, Kahms M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307(5716):1746–1752. doi:10.1126/science.1105654
19. Goodwin JS, Drake KR, Rogers C, et al. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol. 2005;170(2):261–272. doi:10.1083/jcb.200502063
20. Chen S, Zhu B, Yin C, et al. Palmitoylation-dependent activation of MC1R prevents melanomagenesis. Nature. 2017;549(7672):399–403. doi:10.1038/nature23887
21. Hannoush RN, Sun J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat Chem Biol. 2010;6(7):498–506. doi:10.1038/nchembio.388
22. Hernandez JL, Davda D, Majmudar JD, et al. Correlated S-palmitoylation profiling of Snail-induced epithelial to mesenchymal transition. Mol Biosyst. 2016;12(6):1799–1808. doi:10.1039/c6mb00019c
23. Greaves J, Munro KR, Davidson SC, et al. Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry. Proc Natl Acad Sci U S A. 2017;114(8):E1365–E1374. doi:10.1073/pnas.1612254114
24. Hershey JW. The role of eIF3 and its individual subunits in cancer. Biochim Biophys Acta. 2015;1849(7):792–800. doi:10.1016/j.bbagrm.2014.10.005
25. Li W, Li W, Zou L, et al. Membrane targeting of inhibitory Smads through palmitoylation controls TGF-β/BMP signaling. Proc Natl Acad Sci U S A. 2017;114(50):13206–13211. doi:10.1073/pnas.1710540114
26. Hannoush RN, Arenas-Ramirez N. Imaging the lipidome: omega-alkynyl fatty acids for detection and cellular visualization of lipid-modified proteins. ACS Chem Biol. 2009;4(7):581–587. doi:10.1021/cb900085z
27. Liu NQ, Braakman RB, Stingl C, et al. Proteomics pipeline for biomarker discovery of laser capture microdissected breast cancer tissue. J Mammary Gland Biol Neoplasia. 2012;17(2):155–164. doi:10.1007/s10911-012-9252-6
28. Ruggero D. Translation control in cancer etiology. In: Hershey JWB, Sonenberg N, Mathews MB, editors. Protein Synthesis and Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2012:253–279.
29. Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10(4):254–266. doi:10.1038/nrc2824
30. Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi:10.1126/science.1168978
31. Spilka R, Ernst C, Mehta AK, Haybaeck J. Eukaryotic translation initiation factors in cancer development and progression. Cancer Lett. 2013;340(1):9–21. doi:10.1016/j.canlet.2013.06.019
32. Yin Y, Long J, Sun Y, et al. The function and clinical significance of eIF3 in cancer. Gene. 2018;673:130–133. doi:10.1016/j.gene.2018.06.034
33. Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi:10.1146/annurev-biochem-060713-035802
34. Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: newinsights and challenges. In: Hershey JWB, Sonenberg N, Mathews MB, editors. Protein Synthesis and Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2012:29–53.
35. Valásek LS. ‘Ribozoomin‘–translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr Protein Pept Sci. 2012;13(4):305–330.
36. Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500(7462):307–311. doi:10.1038/nature12355
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.