Back to Journals » Neuropsychiatric Disease and Treatment » Volume 13
Analyzing a single nucleotide polymorphism in schizophrenia: a meta-analysis approach
Authors Falola O, Osamor VC, Adebiyi M, Adebiyi E
Received 3 May 2016
Accepted for publication 15 March 2017
Published 23 August 2017 Volume 2017:13 Pages 2243—2250
DOI https://doi.org/10.2147/NDT.S111900
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Roger Pinder
Oluwadamilare Falola,1 Victor Chukwudi Osamor,1,2 Marion Adebiyi,1,2 Ezekiel Adebiyi1,2
1Covenant University Bioinformatics Research (CUBRe), 2Department of Computer and Information Sciences, College of Science and Technology, Covenant University, Ota, Ogun State, Nigeria
Background: Schizophrenia is a severe mental disorder affecting >21 million people worldwide. Some genetic studies reported that single nucleotide polymorphism (SNP) involving variant rs1344706 from the ZNF804A gene in human beings is associated with the risk of schizophrenia in several populations. Similar results tend to conflict with other reports in literature, indicating that no true significant association exists between rs1344706 and schizophrenia. We seek to determine the level of association of this SNP with schizophrenia in the Asian population using more recent genome-wide association study (GWAS) datasets.
Methods: Applying a computational approach with inclusion of more recent GWAS datasets, we conducted a meta-analysis to examine the level of association of SNP rs1344706 and the risk of schizophrenia disorder among the Asian population constituting Chinese, Indonesians, Japanese, Kazakhs and Singaporeans. For a total of 21 genetic studies, including a total of 28,842 cases and 35,630 controls, regression analysis, publication bias, Cochran’s Q and I2 tests were performed. The DerSimonian and Laird random-effects model was used to assess the association of the genetic variant to schizophrenia. Leave-one-out sensitivity analysis was also conducted to determine the influence of each study on the final outcome of the association study.
Results: Our summarized analysis for Asian population revealed a pooled odds ratio of 1.06, 95% confidence interval of 1.01–1.11 and two-tailed P-value of 0.0228. Our test for heterogeneity showed the presence of large heterogeneity (I2=53.44%, P =0.00207) and Egger’s regression test (P =0.8763) and Begg’s test (P =0.8347), indicating no presence of publication bias among our selected studies. In our sensitivity analysis, 10 different studies comprising of ~50% of the entire study had an impact on our final results as each leave-one-out test became insignificant. Our result suggests that genetic variant rs1344706 might be associated with the development of schizophrenia in Asians.
Keywords: meta-analysis, schizophrenia, single nucleotide polymorphism, heterogeneity, genetic variant
Introduction
Schizophrenia is a severe mental disorder, characterized by profound disruptions in thinking, thereby affecting language, perception and sense of self.1 It has been identified as one of the world’s major health disorders. It is one of the disorders that has had a significant effect on the world population, affecting >21 million people. This disorder is mainly common among young people between ages of 15 and 35 years.
The manner of development of schizophrenia may be dependent on our different genetic makeup.2 The genetic differences of individuals from different ethnic groups may determine who is susceptible to schizophrenia or not, and the identification of these variations can be helpful in delivering personalized treatments to patients.3 Personalized treatments also involve the prevention and delivery of treatments to patients, based on their genetic makeup and environment.4 Genome-wide association study (GWAS) has proved to be a useful technique in associating variations in our genome to specific diseases using statistical procedures.
In the event of identifying a significant association between the single nucleotide polymorphism (SNP) and a disease phenotype, such an association is validated by the replication of the statistical procedures on another sample or a smaller subset of the initial sample.5 However, when different authors undertake different GWAS experiments, they sometimes may obtain conflicting reports using the same experimental conditions; conflicts in reports occur when different authors are not able to replicate their results under the same experimental conditions, all things being equal.
Meta-analysis is a technique used to resolve conflicts and discrepancies in GWAS reports. An added advantage of meta-analysis is that it reduces the stress experienced when handling the full genotype data for several GWAS analyses; only the result files are required in a meta-analysis.3,6 Another advantage of meta-analysis in GWAS is the elimination of the dangers of passing on confidential information and genotype data for the cases, as they are not useful in the computation.
Much research has been undertaken to find variations in human DNA that make susceptibility to schizophrenia possible. In the study by O’Donovan et al,7 SNP rs1344706 was identified as a risk factor to the pathogenesis of schizophrenia in European populations. Chen et al8 also found a significant association of SNP rs1344706 with schizophrenia in Chinese population, indicating stronger association of SNP rs1344706 with higher intelligent quotient (IQ) scores in schizophrenia patients. However, Li et al9 did a candidate gene study in two different groups and found no significant association between SNP rs1344706 and schizophrenia in Chinese population. In their analysis, SNP rs359895 was found to have a strong association with schizophrenia in the Chinese population.
Li et al10 did a meta-analysis of 10 Chinese samples and two Japanese samples and found no association with SNP rs1344706. In addition, performing further analysis, they detected that the SNP rs359895 has an association with schizophrenia in the Chinese population. Zhu et al11 performed a meta-analysis containing 13 Chinese samples, 10 Caucasian samples and two samples from other populations. Stratified and combined analysis in the three different groups showed association of SNP rs1344706 with schizophrenia. The abovementioned narrative indicates the presence of inconsistent results in the association of SNP rs1344706 in the Han Chinese population; hence, there is the need of meta-analysis. In this meta-analysis, a total of 21 genetic studies were used, with 13 of these studies reporting no association between the SNP and schizophrenia,7,9,12–19 while eight studies reported that there was an association.8,20–26
Methods
Search for relevant studies from 2006 to 2016 was conducted using online databases which included Web of Science, PubMed and Science Direct. Keywords applied in the search process are listed as “Schizophrenia”, “Chinese”, “association”, “rs1344706” and “ZNF804A” between 2006 and 2016. There were previous meta-analysis studies that were obtained in our initial search, and their bibliographies were searched to obtain more results.
The selection criteria for this meta-analysis included the following: study must be case/control design, study must have at least 100 cases and controls, study must genotype the polymorphism rs1344706 and, finally, the disease being investigated must be schizophrenia. Table 1 shows our initial results.
Description and preprocessing of input data
Table 2 shows the selected studies that were used by the packages for the meta-analysis. The columns include the author name and the year of publication, the number of cases multiplied by 2 (ntrt), the number of controls multiplied by 2 (nctrl), the number of cases that have the risk allele (events in cases, ptrt), the number of controls that have the risk allele (events in controls, pctrl), the frequency of the risk allele (A or T) in affected persons (FA), the frequency of the risk allele in unaffected persons (FU) and the mean ages of cases and controls, and the inference column indicates if the authors’ results show association. The values in ntrt and nctrl columns have been scaled by two compared to that found in Table 1. This is done because an SNP has typically two alleles; therefore, this is accounted for by multiplying the total number of individuals for a study by 2. Table 2 shows the result of preprocessed input data used by our program.
Regression analysis between mean ages and effect sizes
We checked if the mean ages could have influenced the values of the odds ratio (OR). The mean age for each study is shown in Table 1. We could not obtain the mean ages for seven different studies, so we excluded them from our analysis. Our results shown in Table 3 indicate that the OR is not greatly influenced by the mean ages.
  | Table 3 Statistical results of regression analysis between OR and mean age in cases and controls |
From our coefficient of determination (R2), ~3.9% of the OR can be determined by the mean ages for cases and 16% for the controls. In addition, the P-values obtained (P=0.4987) for cases and (P=0.1524) for controls are not statistically significant; hence, the mean ages are not influencing the OR results in any way.
Assessing publication bias
Meta-analysis is a useful technique for finding a true effect size among a distribution of effect sizes. However, the results of a meta-analysis are only as valid as the results of the studies included in it. If studies are selected for inclusion into the meta-analysis with bias, the validity of such analysis will be greatly affected. Publication bias is said to occur when favorable findings are reported by the author rather than unfavorable ones.27,28 In Figure 1, we show the funnel plot for our meta-analysis. The eyeball test shows that the distribution of studies is symmetrical, ie, studies are not coagulated into one quadrant of the funnel.
  | Figure 1 Funnel plot showing publication bias for meta-analysis of rs1344706 (standard error vs log OR). |
The Egger’s regression test (P-value =0.8763) and Begg’s test (P-value =0.8347),29 which are other tools that can also be used to assess publication bias, showed no presence of publication bias.
Assessing degree of heterogeneity
Our Cochran’s Q and I2 tests for heterogeneity showed the presence of large heterogeneity. This is very feasible since most studies were not conducted by the same author, random errors that could occur as a result of mistakes made by the experimenter and actual between variance could be the causes of the amount of heterogeneity observed.
The null hypothesis says that “all studies share a common effect size”, and from our P-value, we reject this hypothesis. The Cochran’s Q and I2 test results shown in Table 4 confirm the fact that all studies do not share a common effect size.
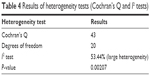  | Table 4 Results of heterogeneity tests (Cochran’s Q and I2 tests) |
Computation of DerSimonian and Laird random-effects model
This meta-analysis used the allelic model of comparison to inspect the association of SNP rs1344706 with schizophrenia in Asian populations, which included Chinese, Kazakhs, Singaporeans, Indonesians and Japanese. Random-effects model was used in the computation of the meta-analysis, since heterogeneity of 54% was obtained. From our results, we observed that individuals who are carriers of the A or T allele of the SNP rs1344706 have a 6% increased risk of susceptibility to schizophrenia in Asian population. Table 5 shows our statistical results; Figure 2 shows the forest plot for included studies.
  | Table 5 Random effect summary results |
Sensitivity analysis
This analysis is performed to check if any single study included in the meta-analysis might have a significant effect on the final results. To achieve this test, data for each study is removed such that the heterogeneity, publication bias, and meta-analysis are then recalculated for the remaining studies, so that the impact of the removed data on the overall study can be ascertained. Table 6 shows the results of our sensitivity analysis. The P-value and OR were not significant for 10 studies of the leave-one-out sensitivity analysis. This indicates that these studies have a significant impact on the results of the final meta-analysis.
Discussion
SNP rs1344706, an intronic SNP in ZNF804A, has been identified with an increased risk of schizophrenia in several human populations. However in the Asian human population, the results have been inconsistent. Steinberg et al14 undertook GWAS and follow-up analysis for several populations and found no significant association between the SNP rs1344706 and schizophrenia in the Chinese population. Li et al9 also did a candidate gene study in two different groups and also found no significant association between SNP rs1344706 and schizophrenia in the Chinese population. In their analysis, SNP rs359895 also located in the same ZNF804A gene was found to have a strong association with schizophrenia in the Chinese population. However, Chen et al,8 investigating the association of SNP rs1344706, found that it had a significant association with schizophrenia in the Chinese population; the interaction of the association with different levels of patient IQ scores was checked, and it was observed that there was a stronger significant association of SNP rs1344706 with higher patient IQ scores. Meta-analysis has been a proven tool useful for resolving conflict reports and to increase power in a study. Meta analysis tries to estimate the consistency of effect size across the group of studies, and based on this consistency, it tries to estimate a summary effect size.30 Meta-analysis was used to find the true effect size of SNP rs1344706 against the risk of schizophrenia.
A total of 21 genetic studies were included in this meta analysis, of which 13 were in support of the null hypothesis, while the other eight studies found a positive association. Looking at the reports of each study that reported no association, reasons for lack of association could be extracted. A common reason for non-association observed from several studies included the risk allele frequencies of the SNP in the comparison groups. Li et al9 argued that studies with allele frequencies that differed to an extent from the frequency of the allele on HapMap31 and from the frequency reported in their studies were likely to have an opposite association. In their work, the T allele frequency of rs1344706 in the comparison (case/control) samples was 0.509, which is similar to the frequencies reported by O’Donovan et al7 (0.514) and Steinberg et al14 (0.546) as well as the frequency in the Han Chinese population from the HapMap31 database (0.524). It is quite different, however, from the frequency reported by Zhang et al21 (0.457) or Chen et al8 (0.564). We performed simple logistic regression to determine if the risk allele frequency of each study might affect the significance of a study result. We obtained a P-value of 0.0596 (~0.06), indicating that the allele frequencies might not be directly influencing the statistical association for each study. Other factors that might affect the association of a genetic study include the use of covariates in the analysis, subgroup analysis performed, disease model or type of statistical analysis used, type of multiple testing corrections used, etc. On the other hand, uncontrolled bias can also affect statistical association. Another strong reason given for the absence of association includes genetic architecture existing between the Asian and Caucasians. Li et al12 stated that the SNP might be inherited together with a functional change in the gene in Europeans, which is not the case in Asians. The linkage disequilibrium map of the genomic region showed a sharp change between the Caucasians and Asians. Wang et al19 stated that demographic differences can be the cause of inconsistencies in results. China is a country known to have complex demography and ethinic diversity, which might cause people from different regions to be born with different genetic traits. Other reasons include genetic heterogeneity, where different alleles are contributing to the pathogenesis of the genetic disorder; environmental exposure; the local culture and difference of schizophrenia subtypes in the subjects included.
The results of this meta-analysis indicated that a statistical significance exists between the phenotype of interest and genotype; several tests were also performed to ensure the validity of the meta-analysis. Regression analysis was performed between the OR and mean ages to check for relationships; publication bias was performed to remove false results, and degree of heterogeneity was checked to detect the variance among the selected studies. This meta-analysis used the allelic model of comparison to inspect the association, and we observed that individuals who are carriers of the A or T allele of the SNP rs1344706 have a 6% increased risk of susceptibility to schizophrenia in the Chinese population.
From the entire population study investigated, the output generated from the meta-analysis portends that there is a significant association between the considered SNP rs1344706 and the prevalence of the schizophrenia disease. However, the systematic investigation of the Chinese population as a subgroup that accounted for 16 studies out a total of 21 studies investigated reveals that there is no significant association between the SNP and the disease condition. The implication may be that the effect of the SNP toward the prevalence of schizophrenia in Chinese is predominantly less compared to other Asian populations considered in this work. Considering the implication of our discovery, the significance of improving and increasing investments in this field of research is recommended by virtue of the fact that the SNP rs1344706 is indicted in the disease risk condition of schizophrenia. This will also help to address the issues of management therapy and treatment discovery for ameliorating the condition of patients plagued with schizophrenia.
Conclusion
Meta-analysis study appears to be a continuous process as fresh genotyping datasets from new GWAS studies for a specific population are made available. To this end, meta-analysis tests are performed to revalidate existing hypotheses using newer GWAS datasets. The association of this SNP has been inspected by four previous meta-analyses,10,11,32,33 with the first study consisting of 12 studies from the Chinese population and stating no association and the second study consisting of 13 studies from the Asian population and stating the presence of association. The third meta-analysis consisted of 30 studies, which included Asians, Caucasians and Americans. The results presented stated the presence of association between the SNP and schizophrenia disorder. The last meta-analysis consisted of 18 studies from Asian population, and the results showed statistical significance. However, we included more datasets in our work for Asian population. Our work also provided a unique perspective view to a combination of population studies from various authors with GWAS experimental data. We also applied R programming, which is a powerful statistical programming tool in our analysis. In addition, Li et al10 had a different result from our analysis, while our result seems to agree with references.11,32,33 It is important to note that due to our interest in understanding the controversy surrounding the association of schizophrenia with SNP rs1344706 among Chinese, Japanese, Singaporean, Indonesian, and Kazakh holistically, it was not in our interest to include populations such as the Caucasian and American as in the study by Sun et al.32
Our result shows that individuals who are carriers of A and T allele of SNP rs1344706 have 6% increased risk of schizophrenia susceptibility among the combined population of Chinese, Japanese, Singaporean, Indonesian and Kazakh. While desiring to investigate the effect of each study on the overall result of the meta-analysis, our sensitivity analysis reveals that 48% of the studies used in the analysis had undue dominance or influence on the overall result. This may be due to other unforeseen factors in the experimental design since we also took account of age and other factors that could bias our results.
The motivation to perform new GWAS studies could discover more polymorphisms associated with the disease as this disease is multigenic or polymorphic with a higher statistical significance; this can be boosted by GWASs with a higher sample size or usage of latest sequencing technologies.
Schizophrenia is one of the severe brain disorders affecting >21 million people in the world today. Although the disorder has a remedy, identifying the areas of the DNA responsible for susceptibility to such a disorder can help in building more therapeutic remedies for it. SNP rs1344706 has been identified as being associated with the susceptibility of schizophrenia in several populations; however, in the Asian and Chinese populations, there have been several conflicting reports. This study thus confirms and validates further the claim by the (eight) previous authors/literature of a significant association between the genotype and phenotype.
Disclosure
The authors report no conflicts of interest in this work.
References
World Health Organization [webpage on the Internet]. Schizophrenia. 2014. Available from: http://www.who.int/mental_health/management/schizophrenia/en/. Accessed April 7, 2015. | ||
Williams HJ, Owen MJ, O’Donovan MC. Schizophrenia genetics: new insights from new approaches. Br Med Bull. 2009;91(1):61–74. | ||
Bush WS, Moore JH. Genome-wide association studies. PLoS Comput Biol. 2012;8(12):e1002822. | ||
Harvey A, Brand A, Holgate ST, et al. The future of technologies for personalized medicine. N Biotechnol. 2012;29(6):625–633. | ||
König IR. Validation in genetic association studies. Brief Bioinform. 2011;12(3):253–258. | ||
Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86(1):6–22. | ||
O’Donovan MC, Craddock N, Norton N, et al; Molecular Genetics of Schizophrenia Collaboration. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40(9):1053–1055. | ||
Chen M, Xu Z, Zhai J, et al. Evidence of IQ-modulated association between ZNF804A gene polymorphism and cognitive function in schizophrenia patients. Neuropsychopharmacology. 2012;37(7):1572–1578. | ||
Li M, Luo XJ, Xiao X, et al. Allelic differences between Han Chinese and Europeans for functional variants in ZNF804A and their association with schizophrenia. Am J Psychiatry. 2011;168(12):1318–1325. | ||
Li M, Zhang H, Luo XJ, et al. Meta-analysis indicates that the European GWAS-identified risk SNP rs1344706 within ZNF804A is not associated with schizophrenia in Han Chinese population. PLoS One. 2013;8(6):e65780. | ||
Zhu M, Liu T, Zhang J, Jia S, Tang W, Luo Y. Association between rs1344706 of ZNF804A and schizophrenia: a meta-analysis. Genomics Proteomics Bioinformatics. 2014;12(6):292–296. | ||
Li M, Shi CJ, Shi YY, et al. ZNF804A and schizophrenia susceptibility in Asian populations. Am J Med Genet B Neuropsychiatr Genet. 2012;159(7):794–802. | ||
Shi Y, Li Z, Xu Q, et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat Genet. 2011;43(12):1224–1227. | ||
Steinberg S, Mors O, Børglum AD, et al. Expanding the range of ZNF804A variants conferring risk of psychosis. Mol Psychiatry. 2011;16(1):59–66. | ||
Liou YJ, Wang HH, Lee MT, et al. Genome-wide association study of treatment refractory schizophrenia in Han Chinese. PLoS One. 2012;7(3):e33598. | ||
Yang Y, Li W, Yang G, et al. Evaluation of the relationship between the ZNF804A single nucleotide polymorphism rs1344706 A/C variant and schizophrenia subtype in Han Chinese patients. Int J Psychiatry Med. 2013;45(3):269–278. | ||
Saito T, Kondo K, Iwayama Y, et al. Replication and cross-phenotype study based upon schizophrenia GWASs data in the Japanese population: support for association of MHC region with psychosis. Am J Med Genet B Neuropsychiatr Genet. 2014;165(5):421–427. | ||
Stepanov VA, Bocharova AV, Saduakassova KZ, et al. Replicative study of susceptibility to childhood-onset schizophrenia in Kazakhs. Russ J Genet. 2015;51(2):185–192. | ||
Wang J, Zhao S, Shugart YY, et al. No association between ZNF804A rs1344706 and schizophrenia in a case-control study of Han Chinese. Neurosci Lett. 2016;618:14–18. | ||
Xiao B, Li W, Zhang H, et al. To the editor: association of ZNF804A polymorphisms with schizophrenia and antipsychotic drug efficacy in a Chinese Han population. Psychiatry Res. 2011;190(2):379–381. | ||
Zhang R, Lu SM, Qiu C, et al. Population-based and family-based association studies of ZNF804A locus and schizophrenia. Mol Psychiatry. 2011;16(4):360–361. | ||
Hashimoto R, Ohi K, Yasuda Y, et al. The impact of a genome-wide supported psychosis variant in the ZNF804A gene on memory function in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2010;153(8):1459–1464. | ||
Schwab SG, Kusumawardhani AA, Dai N, et al; Indonesian Schizophrenia Genetics Consortium. Association of rs1344706 in the ZNF804A gene with schizophrenia in a case/control sample from Indonesia. Schizophr Res. 2013;147(1):46–52. | ||
Kuswanto CN, Woon PS, Zheng XB, et al. Genome-wide supported psychosis risk variant in ZNF804A gene and impact on cortico limbic WM integrity in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2012;159(3):255–262. | ||
Lan X, Wei L, Wu X, et al. Association analysis of ZANF804A rs1344706 polymorphism and schizophrenia. Chin J Nerv Ment Dis. 2013;39(4):213–217. | ||
Zhang C, Wang Z, Hong W, Wu Z, Peng D, Fang Y. ZNF804A genetic variation confers risk to bipolar disorder. Mol Neurobiol. 2015;53(5):2936–2943. | ||
Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Ahmed I, Sutton AJ, Riley RD. Assessment of publication bias, selection bias, and unavailable data in meta-analyses using individual participant data: a database survey. Br Med J. 2012;344:d7762. | ||
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. | ||
Borenstein M, Hedges LV, Higgins J, Rothstein HR. Front Matter. Chichester, England: John Wiley & Sons Ltd; 2009. | ||
International HapMap Consortium. The international HapMap project. Nature. 2003;426(6968):789–796. | ||
Sun Y, Hu D, Liang J, et al. Association between variants of zinc finger genes and psychiatric disorders: systematic review and meta-analysis. Schizophr Res. 2015;162(1):124–137. | ||
Huang L, Ohi K, Chang H, et al. A comprehensive meta-analysis of ZNF804A SNPs in the risk of schizophrenia among Asian populations. Am J Med Genet B Neuropsychiatr Genet. 2016;171B(3):437–446. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




