Back to Journals » Infection and Drug Resistance » Volume 12
Analysis of two pheromone-responsive conjugative multiresistance plasmids carrying the novel mobile optrA locus from Enterococcus faecalis
Authors Shang Y, Li D, Shan X, Schwarz S, Zhang SM, Chen YX, Ouyang W, Du XD
Received 21 February 2019
Accepted for publication 6 June 2019
Published 1 August 2019 Volume 2019:12 Pages 2355—2362
DOI https://doi.org/10.2147/IDR.S206295
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sahil Khanna
Yanhong Shang,1,2,* Dexi Li,2,* Xinxin Shan,2 Stefan Schwarz,3 Su-Mei Zhang,2 Yu-Xia Chen,2 Wuqing Ouyang,1 Xiang-Dang Du2
1Department of Basic Veterinary Medicine, College of Veterinary Medicine, Northwest A&F University, Shaanxi 712100, People’s Republic of China; 2Department of Basic Veterinary Medicine, College of Animal Science and Veterinary Medicine, Henan Agricultural University, Zhengzhou 450046, People’s Republic of China; 3Institute of Microbiology and Epizootics, Centre for Infection Medicine, Department of Veterinary Medicine, Freie Universität Berlin, Berlin, Germany
*These authors contributed equally to this work
Background: The acquired optrA gene, which encodes a ribosomal protection protein of the ABC-F family, can confer cross-resistance to linezolid and florfenicol, posing a serious therapeutic challenge to both human and veterinary medicine.
Purpose: The objective of this study was to investigate the two Enterococcus faecalis (E. faecalis) plasmids for their fine structure, their transferability and the presence of mobile antimicrobial resistance loci.
Methods: To elucidate their fine structure, the two plasmids were completely sequenced and the sequences analysed. Besides conjugation experiments, inverse PCR assays were conducted to see whether minicircles are produced from the mobile antimicrobial resistance loci.
Results: Two pheromone-responsive conjugative optrA-carrying plasmids from E. faecalis, pE211 and pE508 were identified, which can transfer with frequencies of 2.6 ×10−2 and 3.7 ×10−2 (transconjugant per donor), respectively. In both plasmids, optrA was located on the novel mobile optrA locus with different sizes (12,834 bp in pE211 and 7,561 bp in pE508, respectively), flanked by two copies of IS1216 genes in the same orientation. Inverse PCR revealed that circular forms can be generated, consisting of optrA and one copy of IS1216, indicating they are all active. The 77,562 bp plasmid pE211 also carried Tn558 and a mobile bcrABDR locus, and the 84,468 bp plasmid pE508 also harbored the genes fexA, tet(L), tet(O/W/32/O) and a mobile aac(A)-aph(D) locus.
Conclusion: The presence of mobile genetic elements in these plasmids renders them flexible and these elements will aid to the persistence and dissemination of these plasmids among enterococci and potentially also other gram-positive bacteria.
Keywords: enterococci, resistance, IS1216, conjugation, mobile genetic elements
Introduction
Linezolid and florfenicol are both important antimicrobial agents. Linezolid is approved in human medicine and usually used as a last resort antimicrobial agent to treat infections caused by methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE).1 Florfenicol is approved exclusively for food-producing animals, where it is mainly used for the control of respiratory tract infections.2 However, the acquired cross-resistance to linezolid and florfenicol has emerged during the past decade.3–5
Currently, at least three different groups of acquired resistance genes which confer cross-resistance to linezolid and florfenicol have been identified. These include cfr, optrA and poxtA.3–5 Among them, the optrA gene encodes a ribosomal protection protein of the ABC-F family.6 It was first found in Enterococcus faecalis and Enterococcus faecium,4 but has also been detected meanwhile in various Gram-positive bacteria.7,8
In this study, two pheromone-responsive conjugative plasmids harboring optrA along with other resistance genes were analyzed to elucidate the basis for co-selection and dissemination. Furthermore, two novel mobile optrA loci in these plasmids were identified.
Materials and methods
Bacterial strains and antimicrobial susceptibility testing
Two optrA-positive E. faecalis strains (E211–ST59 and E508–ST256) were identified from fecal samples of swine in Henan Province, China during a routine survey in 2015. Antimicrobial susceptibility testing was performed by broth microdilution according to the recommendations given in document M100 (28th ed.) of the Clinical and Laboratory Standards Institute (CLSI).9 S. aureus ATCC 29213 served as the quality control strain.
PCR analysis
The presence of the optrA gene was detected by PCR using the primers listed in Table 1. The optrA-carrying plasmid pE349 was used as the positive control.4 The presence of the circular intermediate was detected by inverse PCR using the primers listed in Table 1. All the PCR products were subjected to Sanger sequencing.
 |
Table 1 PCR primers used in this study |
Transfer experiments
To investigate the transferability of the optrA gene, these two E. faecalis strains were used in conjugation experiments with E. faecalis JH2-2 (rifampicin resistant) as the recipient.10 Transfer frequency is expressed as transconjugant per donor. Colonies that grew on the selective plates supplemented with 50 mg/L rifampin and 10 mg/L florfenicol after incubation for 16–24 h at 37°C were further confirmed by antimicrobial susceptibility testing and multilocus sequence typing (MLST) following harmonized protocols (http://www.mlst.net/).
Sequencing and analysis
Whole genome DNA of two optrA-positive transconjugants E211-T1 and E508-T1 was sequenced by the PacBio RS and Illumina MiSeq platforms. The sequences from PacBio sequencing reads were de-novo assembled and corrected by Illumina MiSeq with pilon. Glimmer 3.02 was used to predict open reading frames (ORFs) and the software blast was used to annotate those ORFs. The sequences determined had been deposited in GenBank under accession numbers MK425644 (pE211) and MK425645 (pE508), respectively.
Results and discussion
The optrA gene in E. faecalis is transferable
The conjugation experiments indicated that these two E. faecalis strains (E211–ST59 and E508–ST256) could transfer florfenicol resistance to the recipient E. faecalis JH2-2 (ST8) at high transfer frequencies, of 2.6×10−2 for E. faecalis E211 and 3.7×10−2 for E. faecalis E508 (transconjugant per donor), respectively. Two transconjugants which were confirmed to share the same background with the recipient (ST8), designated E211-T1 and E508-T1, respectively, were selected for further studies. Sequencing and sequence analysis identified two conjugative optrA-carrying plasmids, designated pE211 and pE508, which were derived from E211-T1 and E508-T1, respectively. The conjugative transfer region in both plasmids pE211 and pE508 displayed the greatest similarity with that in plasmid pTW9, which has key conjugative properties of pheromone-responsive plasmids, such as aggregation substance (As). In combination with with their high transfer frequencies, these two plasmids (pE211 and pE508) can be classified as pheromone-responsive conjugative plasmids. MICs of the two E. faecalis strains, their transconjugants and the recipient strain are shown in Table 2. After transfer, the transconjugants displayed elevated MICs to the respective antimicrobial agents, including florfenicol, linezolid and bacitracin in E211-T1, and florfenicol, linezolid, gentamicin and tetracycline in E508-T1. As shown in Table 3, although there are few amino acid substitutions, the linezolid resistance that the OptrA variants in E. faecalis E211 or E508 confer remains the same with the OptrA prototype in E. faecalis E349.4
 |
Table 2 The antimicrobial susceptibilities of the wild-type strains, transconjugants, and recipient strains in this study |
 |
Table 3 Comparison the difference of OptrA variants and MICs with wide-type strain |
Both plasmids pE211 and pE508 have a novel mobile optrA locus
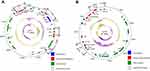
The IS1216-flanked optrA locus in plasmid pE211 consisted of the transcriptional regulator gene araC, the optrA gene and a restriction endonuclease gene (MGE1 in Figure 1A, 12,834 bp), while that in plasmid pE508 carried the optrA gene and a truncated erm(A)-like gene (MGE3 in Figure 1B, 7,561 bp). In both plasmids, optrA was flanked by two copies of IS1216 genes located in the same orientation, forming a novel locus, which was different from that described in previous studies (Figure 2).11,12 In both cases, the two IS1216 elements can recombine and “loop out” a circular intermediate, which can then integrate either into plasmids or in the chromosomal DNA by recombination with another IS1216 copy. Via this way, the optrA gene can move between different chromosomal and plasmidic locations. If finally integrated into a conjugative plasmid or ICE, it can move with this element across strain, species or even genus boundaries. To investigate whether circular intermediates were present, inverse PCR assays using the primers listed in Table 1 were conducted and the results showed that circular intermediates of different sizes (4,052 bp in plasmid pE211 and 3,601 bp in plasmid pE508) were formed in these strains. Sequence analysis of these circular intermediates confirmed that they contained one copy of the IS1216 element and the sequence that was formerly located between the two IS1216 elements, including optrA.
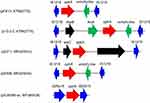 |
Figure 2 The environment of the optrA gene in different plasmids. |
The IS1216-like elements have also been reported to be associated with the vancomycin resistance VanA gene cluster in E. faecium,13 the multidrug resistance genes poxtA, optrA and cfr in enterococci and staphylococci,5,11,14 the macrolide-lincosamide-streptogramin B resistance genes erm(B) and erm(T) in enterococci and streptococci,15,16 and the tetracycline resistance gene tet(S) in Streptococcus infantis.17 These observations, along with multiple MGEs (MGE1-MGE4) associating with IS1216 elements found in this study, suggest that the IS1216-like elements could play an important role in dissemination of the respective antimicrobial resistance genes among various Gram-positive organisms.
The analysis of the genetic context of optrA in plasmids pE211 and pE508
As shown in Figure 1A and Table 4, the 77,562 bp plasmid pE211 harbored the phenicol/oxazolidinone resistance gene optrA, the TN558- associated phenicol resistance gene fexA, and the mobile bacitracin resistance operon bcrABDR (Mobile Genetic Element, MGE2, 5,527 bp). The fexA-carrying transposon Tn558 has previously been described on plasmids in Staphylococcus lentus, Staphylococcus cohnii, and Enterococcus spp.11,18,19 Here, it is present on plasmid pE211 in E. faecalis. The MGE2 consisting of the bcrABDR operon confers resistance to bacitracin. The bcr locus was flanked by ISEnfa1 elements as previously described in E. faecalis or Clostridium perfringens.20,21 Here, it is present on the plasmid pE211 in E. faecalis.
 |
Table 4 Coding sequences of the plasmid pE211 |
As shown in Figure 1B and Table 5, the 84,468 bp plasmid pE508 harbored the phenicol/oxazolidinone resistance gene optrA, the phenicol resistance gene fexA, the mobile bifunctional aminoglycoside resistance gene aac(A)-aph(D) locus (MGE4, 5,891 bp), and the tetracycline resistance genes tet(L) and tet(O/W/32/O). The aminoglycoside resistance gene aac(A)-aph(D) is usually located on the transposon Tn4001 from staphylococci, Tn5281 from enterococci or Tn3706 from streptococci. Together with other resistance genes, it can also be located on the transposons Tn924, Tn5384 or Tn5385 from E. faecalis.22 In this study, to the best of our knowledge, it was for the first time seen that aac(A)-aph(D) is flanked by two copies of IS1216 elements located in the same orientation on the plasmid pE508 from E. faecalis.
 |
Table 5 Coding sequences of plasmid pE508 |
The presence of the circular intermediates in MGE2 and MGE4 were detected by inverse PCR (Table 2) and further sequence analysis indicated that both MGEs are active. However, the Tn558 locus is apparently not active as no circular intermediates were detectable.
Conclusion
Two pheromone-responsive conjugative multiresistance plasmids carrying the novel optrA locus from E. faecalis were identified, with one plasmid (pE211) harbouring a mobile bcrABDR locus, and the other (pE508) a mobile aac(A)-aph(D) locus. All these mobile locus were active due to the presence of the minicircles. The presence of MGEs in these plasmids renders them flexible and these elements will aid to the persistence and dissemination of these plasmids among enterococci and potentially also other Gram-positive bacteria.
Acknowledgments
We thank Don B. Clewell (University of Michigan) for providing E. faecalis reference strain JH2-2. This work was supported by grants from the National Natural Science Foundation of China (No. U1604103, U1704108 and 31472239), the Program for Innovative Research Team (in Science and Technology) in University of Henan Province (No. 18IRTSTHN020) and the German Federal Ministry of Education and Research (BMBF) under project number 01KI1727D as part of the Research Network Zoonotic Infectious Diseases.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hashemian SMR, Farhadi T, Ganjparvar M. Linezolid: a review of its properties, function, and use in critical care. Drug Des Devel Ther. 2018;12:1759–1767. doi:10.2147/DDDT.S164515
2. El Garch F, de Jong A, Simjee S, et al. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe, 2009-2012: VetPath results. Vet Microbiol. 2016;194:11–22. doi:10.1016/j.vetmic.2016.04.009
3. Schwarz S, Werckenthin C, Kehrenberg C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother. 2000;44:2530–2533. doi:10.1128/aac.44.9.2530-2533.2000
4. Wang Y, Lv Y, Cai J, et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother. 2015;70:2182–2190. doi:10.1093/jac/dkv116
5. Antonelli A, D’Andrea MM, Brenciani A, et al. Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J Antimicrob Chemother. 2018;73:1763–1769. doi:10.1093/jac/dky088
6. Sharkey LK, Edwards TA, O’Neill AJ. ABC-F proteins mediate antibiotic resistance through ribosomal protection. MBio. 2016;7:e01975. doi:10.1128/mBio.01975-15
7. Fan R, Li D, Feßler AT, Wu C, Schwarz S, Wang Y. Distribution of optrA and cfr in florfenicol-resistant Staphylococcus sciuri of pig origin. Vet Microbiol. 2017;210:43–48. doi:10.1016/j.vetmic.2017.07.030
8. Huang J, Chen L, Wu Z, Wang L. Retrospective analysis of genome sequences revealed the wide dissemination of optrA in Gram-positive bacteria. J Antimicrob Chemother. 2017;72:614–616. doi:10.1093/jac/dkw488
9. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
10. Huys G, D’Haene K, Collard JM, et al. Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl Environ Microb. 2004;70:1555–1562. doi:10.1128/AEM.70.3.1555-1562.2004
11. He T, Shen Y, Schwarz S, et al. Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J Antimicrob Chemother. 2016;71:1466–1473. doi:10.1093/jac/dkw016
12. Morroni G, Brenciani A, Antonelli A, et al. Characterization of a multiresistance plasmid carrying the optrA and cfr resistance genes from an Enterococcus faecium clinical isolate. Front Microbiol. 2018;9:2189. doi:10.3389/fmicb.2018.02189
13. Darini AL, Palepou MF, Woodford N. Effects of the movement of insertion sequences on the structure of VanA glycopeptide resistance elements in Enterococcus faecium. Antimicrob Agents Chemother. 2000;44:1362–1364. doi:10.1128/aac.44.5.1362-1364.2000
14. Liu Y, Wang Y, Schwarz S, et al. Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment. Antimicrob Agents Chemother. 2013;57:42–48. doi:10.1128/AAC.01605-12
15. Raze D, Dardenne O, Hallut S, Martinez-Bueno M, Coyette J, Ghuysen JM. The gene encoding the low-affinity penicillin-binding protein 3r in Enterococcus hirae S185R is borne on a plasmid carrying other antibiotic resistance determinants. Antimicrob Agents Chemother. 1998;42:534–539.
16. Tsai JC, Hsueh PR, Chen HJ, Tseng S-P, Chen P-Y, Teng L-J. The erm(T) gene is flanked by IS1216V in inducible erythromycin-resistant Streptococcus gallolyticus subsp. pasteurianus. Antimicrob Agents Chemother. 2005;49:4347–4350. doi:10.1128/AAC.49.10.4347-4350.2005
17. Ciric L, Brouwer MS, Mullany P, Roberts AP. Minocycline resistance in an oral Streptococcus infantis isolate is encoded by tet(S) on a novel small, low copy number plasmid. FEMS Microbiol Lett. 2014;353:106–115. doi:10.1111/1574-6968.12410
18. Kehrenberg C, Schwarz S. Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob Agents Chemother. 2005;49:813–815. doi:10.1128/AAC.49.2.813-815.2005
19. Wang Y, Zhang W, Wang J, et al. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob Agents Chemother. 2012;56:1485–1490. doi:10.1128/AAC.05827-11
20. Wang XM, Li XS, Wang YB, et al. Characterization of a multidrug resistance plasmid from Enterococcus faecium that harbours a mobilized bcrABDR locus. J Antimicrob Chemother. 2015;70:609–611. doi:10.1093/jac/dku416
21. Charlebois A, Jalbert LA, Harel J, Masson L, Archambault M, Tse H. Characterization of genes encoding for acquired bacitracin resistance in Clostridium perfringens. PLoS One. 2012;7:e44449. doi:10.1371/journal.pone.0044449
22. Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect. 2010;16:541–554. doi:10.1111/j.1469-0691.2010.03226.x
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.