Back to Journals » Cancer Management and Research » Volume 11
Analysis of the circular RNA transcriptome in the grade 3 endometrial cancer
Authors Ye F, Tang QL, Ma F, Cai L, Chen M, Ran XX, Wang XY, Jiang XF
Received 6 December 2018
Accepted for publication 29 May 2019
Published 5 July 2019 Volume 2019:11 Pages 6215—6227
DOI https://doi.org/10.2147/CMAR.S197343
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Rituraj Purohit
Fei Ye1,*, Qiong Lan Tang2,*, Fang Ma1,*, Lei Cai,1 Ming Chen,1 Xiao Xia Ran,1 Xiao Yu Wang,1 Xue Feng Jiang1
1Department of Obstetrics and Gynecology, The First Affiliated Hospital, Jinan University, Guangzhou 510630, People’s Republic of China; 2Department of Pathology, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou 510120, People’s Republic of China
*These authors contributed equally to this work
Background/aims: Circular RNAs (circRNAs), a class of newly discovered endogenous noncoding RNAs, have shown large capabilities in gene regulation. Patients with the grade 3 endometrial cancer (EC) have a generally poor prognosis, and the specific role of circRNAs in the grade 3 EC remains unclear. This study aims to investigate the roles of circRNAs in the grade 3 EC.
Methods: In the current study, we screened the expression profiles of circRNAs taken from two women with the grade 3 EC and adjacent non-cancerous endometrial tissue using circRNAs sequencing. Bioinformatic analyses were applied to study these differentially expressed circRNAs. Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) of six dysregulated circRNAs was performed to validate the sequencing results. Bioinformatic analyses, including the negative correlation network analyses of circRNAs–microRNAs (miRNAs)–messenger RNAs (mRNAs) and the Cytoscape, were used to delineate the interaction of circRNAs/miRNAs of the entire network.
Results: Data of circRNA sequencing showed a significant change in 75,928 unique circRNAs (P<0.05). The upregulated hsa_circ_0039569 and hsa_circ_0001610 and downregulated hsa_circ_0000437, hsa_circ_0001776, and hsa_circ_0009043 were validated by qRT-PCR analysis. Using bioinformatical methods, we found that hsa_circ_0039569 has the MRE of hsa-miR-542-3p and hsa-let-7c-5p. Hsa-miR-542-3p and hsa-let-7c-5p were downregulated in the grade 3 EC validated by qRT-PCR analysis. In the clinicopathological parameters, the expression level of hsa_circ_0039569 was significantly correlated with tumor differentiation (P=0.001).
Conclusion: This is the first study demonstrated that there were a lot of differences between the tissue of the grade 3 EC and adjacent non-cancerous endometrial in circRNA expression and may offer novel molecular candidates for diagnosis and clinical treatment of the grade 3 EC.
Keywords: grade 3 endometrial cancer, circRNAs, transcriptome, RNA-Seq
Introduction
Endometrial cancer (EC) is the most common gynecological malignancy of the female reproductive system. Traditionally, EC was divided into type 1 and type 2,1 grades 1 and 2 are considered “type 1” EC, while grade 3 is considered “type 2”. Type 1 may arise from atypical hyperplasia and is associated with excess progesterone and estrogen receptor. In addition, the prognosis is relatively good. Type 2 may arise from atrophic endometrium and is associated with a lack of estrogen and progesterone receptor.However, it shows the worse prognosis than type 1 because of the aggressive cancerous tissue.
Non-coding RNAs (ncRNAs) play an important role in epigenetics increasingly. NcRNAs can be divided into circular non-coding RNAs (circRNAs) and linear ncRNAs. However, in recent years, we have observed the explosive growth of published research on various aspects of circRNA biology. It indicates that this molecular plays an important role in normal cellular differentiation, tissue homeostasis as well as in the development of the disease. As it is known that circRNAs arise from precursor mRNAs, their biogenesis remains unspecific. circRNAs differ from other RNAs in their extraordinary continuous closed loop structure, which is covalently linked by free 3′ and 5′ ends.2 It is also called a “back-splicing” structure,3 and is generated from the joining of an upstream 3′ splice site to a downstream 5′ splice site, which prevents them from degradation by RNA exonucleases.4 There are many biological functions of circRNA, and the most important function is to regulate gene expression. An increasing number of studies have found that many circRNAs as miRNA sponges or competitive endogenous RNAs (ceRNAs) can regulate the expression of miRNA targets, hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via sponging miR-492,5 hsa_circZNF609 promotes breast cancer cell growth, migration, and invasion via sponging miR-145-5p.6 In addition to acting as miRNA sponges, circRNAs can also regulate the transcription and alternative splicing. The experiment of co-localization indicated that circRNAs could interact with RBPs. It has been reported the interaction among circ-Foxo3 with E2F1, ID-1, HIFα, and FAKs played a part in anti-aging and anti-stress.7 Besides the non-coding function of circRNAs, scientists have also studied the capacity of translation of circRNAs.8 Based upon the functions of circRNAs, researchers have studied the role of circRNAs in pathology and physiology. The evidence show that circRNAs exert a regulatory function in different diseases via different mechanisms. Moreover, circRNAs have the potential to serve as clinical diagnosis markers and therapeutic targets.9
To date, only one study has shown that circRNAs may play roles in EC, with grade (1–2) type I EC.10 However, it was known little about the underlying regulatory mechanisms of circRNAs with the grade 3 EC. To our knowledge, this was the first study of circRNAs transcriptome analysis of the grade 3 EC in the worldwide. We made a description about the factor of circRNAs transcriptome analysis of the grade 3 EC, detected the differentially expressed circRNAs, and made a further analysis of the related microRNAs of the differentially expressed circRNAs. In short, the differentially expressed circRNAs might become the diagnosis, treatment, and the potential biomarker of the prognosis of the grade 3 EC.
Materials and methods
Ethics statement
The project was approved and supervised by the Ethics Committee of the First Affiliated Hospital of Jinan University, Guangzhou, People's Republic of China. All samples were collected from the patient who had undergone surgery in the First Affiliated Hospital of Jinan University. All the enrolled patients have signed the informed consent. This study was conducted in accordance with the Declaration of Helsinki.
EC and normal tissue samples
Tissues were collected from the patients with EC undergone surgery at the Department of Surgery at the First Affiliated Hospital of Jinan University, and the nontumorous tissue was collected at the site 10 mm away from the affected region at the same time. The tumor specimen was staged and classified by pathological result. Two pairs of snap-frozen endometrial tissue with the grade 3 EC and adjacent non-cancerous endometrial tissue were collected for circRNA sequencing analysis (Tables 1 and 2) and another 60 samples (15 pairs of the grade 3 EC and adjacent non-cancerous endometrial tissue, 15 pairs of the grade 1–2 EC and adjacent non-cancerous endometrial tissue) were collected for qRT-PCR validation.
 |
Table 1 Patient and tissue sample chart |
 |
Table 2 Detailed clinical pathological parameters of patients being sequenced |
RNA isolation
We used TRIzol (Invitrogen, Carlsbad, CA, USA) to extract total RNA from each frozen specimen and treating them with DNase (Takara, Dalian, People's Republic of China) according to the manufacturer’s instructions. The RNA integrity was assessed using an Agilent Bioanalyzer2100 (Agilent Technologies, Santa Clara, CA, USA). The quantity (ng/mL) and purity (260/280 and 260/230 ratios) of total RNAs were assessed using NanoDrop 2,000 spectrophotometer (Thermo, Waltham, MA, USA) and denaturing agarose gel electrophoresis. Each RNA sample had an A260:A280 ratio above 1.9 and an A260:A230 ratio above 1.8. The values of RIN of A1, A2, B1, and B2 were 8.8, 8.7, 8.9, and 8.6.
Circrnas sequencing analysis
RNA library preparation and circRNA sequencing were performed by the Beijing Genomics Institute (Shengzheng, People's Republic of China). The sample preparation and microarray hybridization were performed according to the Arraystar’s standard instructions. In short, total RNA samples were digested with RNase R (Epicenter Inc, San Francisco, CA, USA) to remove linear RNAs and enrich circRNAs. The rRNA-depleted RNA was used to build up the RNA libraries with TruSeq Stranded Total RNA Library Prep Kit (Illumina, San Diego, CA, USA). Quality of libraries were checked with BioAnalyzer 2100 system (Agilent Technologies, Richardson, TX, USA). The RNA libraries were denatured as single-stranded DNA molecules. The cDNAs were captured on Illumina Flow Cells (Illumina, San Diego, CA, USA) amplified in situ as clusters and finally sequenced with 150-bp paired reads on HiSeq 4000 sequencing system (Illumina, San Diego, CA, USA).
We used Cluster and Tree View software to perform hierarchical clustering analysis, in order to analyze the differentially expressed circRNAs between the grade 3 EC and adjacent non-cancerous endometrial tissue. The predicted functions of the differentially expressed circRNAs between the grade 3 EC and adjacent non-cancerous endometrial tissue were obtained by GO (http:\\www.geneontology.org\) and KEGG pathway analysis (http:\\www.genome.jp\kegg\).
Delineation of circrna/mirna interactions
The circRNA/microRNA interaction was predicted using Arraystar’s home-made miRNA target prediction software based on TargetScan and miRanda.11,12 We searched MREs on circRNAs using the software and then selected the miRNAs according to seed-matching sequence to establish circRNA-miRNA network. Then, we used Cytoscape (version 3.6.0, USA) to draw the graph of the circRNA/miRNA network.
Quantitative real‐time polymerase chain reaction (qRT‐PCR) validation
MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) was used for synthesizing cDNA according to manufacturer’s instructions after RNA extraction. The expression level of the circRNAs was evaluated by SYBR Green qPCR. Specific divergent primers were designed to amplify the circular transcripts, only primers achieving a single peak in the melting curve were considered proper for qRT-PCR validation. PCR was performed in 10 μL reaction volume, including 2 μL of cDNA, 5 μL 2×Master Mix, 0.5 μL of Forward Primer (10 μM), 0.5 μL of Reverse Primer (10 μM), and 2 μL of double-distilled water. The reaction was set at 95°C for 10 mins for pre-denaturation, then at 95°C for 10 s and at 60°C for 60 s repeating 40 cycles. Housekeeping gene U6 was used as an internal control. Both target and reference were amplified in triplicate wells. The relative expression of candidate circRNAs was normalized to U6 and then calculated using the 2−ΔΔCt method.Primers are designed using PRIMER5 software (Table 3).
 |
Table 3 Primers allowed for amplification of genes in this work |
Statistical analysis
All statistical analyses in this study were accomplished by the GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA) and the SPSS 23.0 software package (IBM, Chicago, IL, USA). Paired t test, independent t test, and chi-square test were used in this study accurately. P-value of 0.05 or less was considered statistically significant.
Results
Identification of differentially expressed circrna profiles
High throughput sequencing analysis facilitated our research on circRNA expression signatures in the grade 3 EC. A total of 75,928 circRNA targets were detected by microarray probes in two pairs of samples. In the initial stage of experiments, circRNA expression profile of two pairs of the grade 3 EC and adjacent non-cancerous tissues were performed. We confirmed the differentially expressed circRNA with statistical significance between the two groups by using Scatter plot filtering and Volcano Plot filtering (Figure 1A and B). Box plots show the normalized intensities from the two groups (Figure 1C). What is more, we analyzed the category of differentially expressed circRNAs. Most of them are transcribed from the introgenic, some are from intergenic (Figure 1D). Furthermore, we analyzed the distribution of the differential expression of circRNAs on all the human chromosomes (Figure 2).
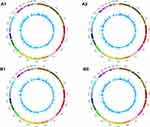 |
Figure 2 The distribution of differentially expressed circRNAs in human chromosomes (A1, A2, B1, and B2). |
Identification of differentially expressed circRNAs in patients with the grade 3 EC and adjacent non-cancerous endometrial tissue
Hierarchical cluster was performed to analyze all the circRNAs and shown the top 10 up and downregulated circRNAs between the grade 3 EC and adjacent non-cancerous endometrial tissues (Figure 3A and B). The data suggested that the expression of circRNAs in the grade 3 EC tissues was different from the matched non-tumor tissue. A total of 62,167 unique circRNAs were significantly altered in women with the grade 3 EC and adjacent non-cancerous endometrial tissue (P<0.05). Among them, 25,735 were significantly upregulated and 36,432 were down-regulated. The top 10 up and downregulated circRNAs between the two groups are listed in Tables 4 and 5, respectively. Among these, the expression levels of hsa_circ_0039569, hsa_circ_0001523, hsa_circ_0001610, hsa_circ_0001400, and hsa_circ_0007905 were up-regulated, and hsa_circ_0000437, hsa_circ_0001776, hsa_circ_0009043, hsa_circ_0000471, and hsa_circ_0014606 were downregulated as top 5, respectively.
 |
Table 4 Comparison of the grade 3 EC with the adjacent non-cancerous endometrial tissue for the top 10 upregulated circRNAs (fold change>2 and P<0.05) ranked by fold changes in microarray date |
 |
Table 5 Comparison of the grade 3 EC versus the adjacent non-cancerous endometrial tissue for the top 10 downregulated circRNAs (fold change>2 and P<0.05) ranked by fold changes in microarray data |
Construction of the circRNA/miRNA interaction network and prediction of circRNA-miRNA associations
Considered that the circRNA had the potential to serve as miRNA sponges to regulate gene expression, so we made a further exploration about the function of ceRNA of circRNA. We used TargetScan and miRanda database software to predict the target miRNAs of circRNAs based on the base pairing rule in Arraystar. All the differentially expressed circRNAs were predicted according to the complementary miRNA matching sequence. There were a total of 451 miRNAs, the top 10 upregulated and downregulated circRNAs, could be combined with circRNAs. Then, used Cytoscape delineated an entire network of circRNA/miRNA interaction (Figure 4).
Validation for the differential expression level of circRNAs
To confirm the data analysis expression of circRNA microarray, six circRNAs (hsa_circ_0039569, hsa_circ_0001523, hsa_circ_0001610, hsa_circ_0000437, hsa_circ_0001776, and hsa_circ_0009043) were validated out using qRT-PCR in 30 pairs of samples including 15 pairs of the grade 3 EC and adjacent non-cancerous endometrial tissue, 15 pairs of the grade 1–2 EC and adjacent non-cancerous endometrial tissue. The results showed that circRNAs were significantly differentially expressed between the grade 3 EC and the grade 1–2 EC.
The qRT‐PCR results are presented in Figure 5. It showed that the expression of the circRNAs hsa_circ_0039569 and hsa_circ_0001610 were upregulated at the grade 3 EC and the grade 1–2 EC tissue versus the adjacent non-cancerous endometrial tissue, compared with the grade 1–2 EC tissue, hsa_circ_0039569 and hsa_circ_0001610 have a higher level of expression in the grade 3 EC. The hsa_circ_0001523 qPCR results were inconsistent with the microarray analysis. The hsa_circ_0009043, hsa_circ_0000437, and hsa_circ_0001776 were significantly downregulated at the grade 3 EC and the grade 1–2 EC tissue versus the adjacent non-cancerous endometrial tissue. And the hsa_circ_0009043 and hsa_circ_0001776 had a higher level of expression in the grade 3 non-cancerous endometrial tissue versus the grade 1–2 non-cancerous endometrial tissue, whereas hsa_circ_0000437 had a lower level of expression.
The relationship between the expression of hsa_circ_0039569 and clinicopathological parameters of the 30 patients
The qPCR results showed that hsa_circ_0039569 was higher expression of the grade 3 EC versus the grade 1–2 EC. To analyze the relationship between the expression of hsa_circ_0039569 and clinicopathological parameters of the 30 patients, the patients were divided into low expression group (n=15) and high expression group (n=15) with the hsa_circ_0039569 relative expression level 2−ΔΔCт, median is 2.985. The correlation between the hsa_circ_0039569 and clinicopathological data was analyzed. The results showed that the expression level of hsa_circ_0039569 was not correlated with age, lymph node metastasis, tumor size, FIGO stage, and myometrial invasion (P>0.05), and was significantly correlated with tumor differentiation (P=0.001). The difference was statistically significant (Table 6).
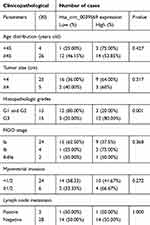 |
Table 6 The relationship between the expression of hsa_circ_0039569 and clinicopathological parameters of the thirty patients |
Prediction of circRNA-miRNA associations
In order to explore the mechanism of circRNAs, the circRNA-miRNA axis in cancer-related pathways was predicted. DIANA-miRPath determined all the candidate miRNAs that were involved in possible pathways by p-value cutoff at 0.05. The data showed that hsa_circ_0039569 was associated with cancer-related pathways for the grade 3 EC with the greatest potential. Base on TargetScan and miRanda, the hsa_circ_0039569/microRNAs interaction was predicted using Arraystar’s home-made miRNA target prediction software. KEGG analysis of all microRNAs showed that miR-542-3p and -let-7c-5 was associated with cancer-related pathways for the grade 3 EC with the greatest potential. Firstly, we further analyzed the binding sequence between hsa_circ_0039569 and hsa-miR-542-3p or hsa-let-7c-5p interaction by TargetScan and miRanda analysis. We found circ_0039569 had a perfect match sequence (7-mer-m8 seed type) to bind miR-542-3p (Figure 6A) or let-7c-5p (Figure 6B). To made a confirmation of the relationship between circRNA between hsa_circ_0039569 and hsa-miR-542-3p or hsa-let-7c-5p, hsa-miR-542-3p and hsa-let-7c-5p were validated by using qRT-PCR in 30 pairs of samples.
The qRT‐PCR results are presented in Figure 7. It showed that the expression of the microRNAs hsa-miR-542-3p and hsa-let-7c-5p was upregulated at the grade 3 non-cancerous endometrial tissue and the grade 1–2 non-cancerous endometrial tissue versus the the grade1-3 EC tissue, compared with the grade 1–2 non-cancerous endometrial tissue, hsa-miR-542-3p, and hsa-let-7c-5p have a higher level of expression in the non-cancerous endometrial tissue. So, the hsa_circ_0039569-hsa-miR-542-3p/hsa-let-7c-5p axis was predicted as one of the circRNA-miRNA axis. Spearman correlation test was used to analyze the correlation between hsa_circ_003956 and hsa-miR-542-3p and hsa-let-7c-5p in the grade 3 EC. The correlation coefficient R results are -8923 and -0.9027 (Figure 8). So, hsa_circ_003956 has a negative correlation with hsa-miR-542-3p and hsa-let-7c-5p.
 |
Figure 8 The result of spearman correlation test between hsa_circ_003956 and hsa-miR-542-3p/hsa-let-7c-5p in the grade 3 EC. |
Discussion
Presently, the mechanism and role of circRNAs in cancer progression haven’t been well identified.13 Emerging evidence have found that circRNAs played an important part role in multiple pathological and physiological functions. The available evidence showed that circRNAs are associated with proliferation,14 cell cycle,15 aoptosis,16 and autophagy,17 involving nervous system disease, cardiovascular system disease, and series of tumors.18,19 Especially, more and more studies have found that circular RNA plays an increasingly important role in malignant tumors which were related to sex hormones. Accumulating evidence indicates that hsa_circ_006054, hsa_circ_100219 hsa_circ_0006528, and circ-ABCD10 might also play roles in breast cancer.20–22 Furthermore, hsa_circ_0001982 promotes breast cancer cell carcinogenesis by targeting miR-143.23 Additionally, the expression of circ-Foxo3 was downregulated in mouse breast tumor tissues compared with normal breast, lipid tissues, and skin, and as an inhibitor of breast cancer tumor cells inhibits tumor cell proliferation and survival.24 It has been proved that circ-SMARCA5 acts as an oncogene in prostate cancer by promoting the cell cycle and inhibiting apoptosis.25
In the field of EC, up to the present time, just one study has shown that circRNAs may play roles in EC, with grade (1–2) type I EC, it reported the circular transcriptome specific for EC as determined by RNA sequencing and found that the abundance of circRNAs was lower in the grade 1–2 EC than in adjacent non-cancerous endometrial tissue. What is more, they found that numerous “hotspot” genes from which circRNAs were transferred that may account for alterations in circRNA expression between the non-cancerous endometrial tissue and the grade 1–2 EC. Most importantly, they have also identified circRNAs which were differentially expressed between the grade 1–2 EC and normal endometrial tissue. More and more researchers acknowledge that the patients with the grade 3 EC have a generally poor prognosis,26 and the specific role of circRNAs in the grade 3 EC remains unclear.
In this study, the differentially expressed profiles of circRNAs in the grade 3 EC tissue and adjacent non-cancerous endometrial tissue were analyzed and validated. Up to now, this is the first study to evaluate the differential expression of circRNAs in the grade 3 EC patients. It indicated that the grade 3 EC-associated circRNAs could be exploited as new candidates for diagnosis and treatment of the grade 3 EC.
Base on the above study, there were 25,735 circRNAs significantly upregulated and 36,432 circRNAs significantly downregulated in the grade 3 EC tissue compared with adjacent non-cancerous endometrial tissue. Among these, the expression levels of hsa_circ_0039569, hsa_circ_0001523, hsa_circ_0001610, hsa_circ_0001400, and hsa_circ_0007905 were upregulated, and hsa_circ_0000437, hsa_circ_0001776, hsa_circ_0009043, hsa_circ_0000471, and hsa_circ_0014606 were downregulated as top 5, respectively. The upregulated hsa_circ_0039569 and hsa_circ_0001610 and downregulated hsa_circ_0000437, hsa_circ_0001776, and hsa_circ_0009043 were validated by qRT-PCR analysis. Hsa_circ_0039569 were selected for further analysis, in the clinicopathological parameters of the 30 patients, the expression level of hsa_circ_0039569 was significantly correlated with tumor differentiation (P=0.001), and was not correlated with age, lymph node metastasis, tumor size, FIGO stage and myometrial invasion (P>0.05). Furthermore, using Cytoscape to delineate an entire network of circRNAs/miRNAs interaction. This was the first time of our study which predicted hundreds of circRNAs/miRNAs interaction. An interaction network of circRNAs/miRNAs was constructed, which provided valuable information for ceRNA research of circRNAs in the grade 3 EC. In addition, the results showed that the hsa_circ_0039569 was highly expressed in the grade 3 EC compared to control. With bioinformatical methods, we found that hsa_circ_0039569 had MREs of hsa-miR-542-3p and hsa-let-7c-5p. Hsa-miR-542-3p and hsa-let-7c-5p were downregulated at the grade 3 EC were validated by qRT-PCR analysis. Predicting that hsa_circ_0039569-hsa-miR-542-3p/hsa-let-7c-5p was one of the circRNA-miRNA axis. Our results indicated that it was worthwhile to investigate these novel dysregulated circRNAs as miRNA sponges and their potential biological functions in the grade 3 EC.
All in all, for the first time, our study revealed the circRNA expression signatures of the grade 3 EC. Furthermore, the regulation of hsa_circ_0039569-hsa-miR-542-3p/hsa-let-7c-5p axis was initially predicted in the grade 3 EC. Next, our laboratory is doing further research on the mechanism of hsa_circ_0039569. The result of our study emphasized that hsa_circ_0039569 could be an important prediction of the grade 3 EC. This study has laid the foundation for the functional studies, further diagnosis, treatment of circRNAs of the grade 3 EC.
Conclusion
This study demonstrated that lots of differentially expressed cricRNAs in endometrial tissue of the grade 3 EC patients compared with adjacent non-cancerous endometrial tissue. Therefore, it may offer novel molecular candidates for diagnosis and clinical treatment of the grade 3 EC.
Acknowledgments
We thank all the donors who participated in the work. This study was supported by the Natural Science Foundation of Guangdong Province (No: 2016A030313115).
Author contributions
Fei Ye, Xiao Yu Wang, and Xue Feng Jiang conceived and designed the experiments. Fang Ma and Qiong Lan Tang performed the experiments. Lei Cai, Ming Chen, and Xiao Xia Ran analyzed the data. Fei Ye prepared the figures and wrote the manuscript. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15(1):10–17.
2. Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta Gene Regul Mech. 2016;1859(10):1245–1251. doi:10.1016/j.bbagrm.2016.07.009
3. Barrett SP, Salzman J. Circular RNAs:analysis, expression and potential functions. Development. 2016;143(11):1838–1847. doi:10.1242/dev.128074
4. Qu S, Yang X, Li X, et al. Circular RNA:A new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
5. Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag Res. 2019;11:1033–1041. doi:10.2147/CMAR.S186857
6. Wang S, Xue X, Wang R, et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manag Res. 2018;10:3881–3890. doi:10.2147/CMAR.S174778
7. Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:402–1412.
8. Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37. doi:10.1016/j.molcel.2017.02.017
9. Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacology&Therapeutics. 2018;187:31–44.
10. Chen BJ, Frances L, Byrne F, et al. Analysis of the circular RNA transcriptome in endometrial cancer. Oncotarget. 2018;9(5):5786–5796.
11. Pasquinelli A. MicroRNAs and their targets: recognition,regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. doi:10.1038/nrg3162
12. Enright AJ, John B, Gaul U, Tuschl, T., Sander, C., Marks, D S. MicroRNA targets in drosophila. Genome Biol. 2003;5:R1. doi:10.1186/gb-2003-5-1-r1
13. Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–1045. doi: 10.1080/15476286
14. Bachmayr-Heyda A, Reiner AT, Auer K, et al. PilsCorrelation of circular RNA abundance with proliferation—exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5(1):8057. doi:10.1038/srep08057
15. Du WW, Yang W, Liu E, Yang, Z., Dhaliwal, P., Yang, B B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi:10.1093/nar/gkw027
16. Du WW, Fang L, Yang W, Wu, N., Awan, F M., Yang, Z., Yang, B B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi:10.1038/cdd.2016.133
17. Huang R, Zhang Y, Han B, et al Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13(10):1722–1741. doi:10.1080/15548627.2017.1356975
18. Greene J, Baird AM, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi:10.3389/fmolb.2017.00038
19. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116:626–633. doi:10.1038/bjc.2016.451
20. Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9:1175–1188. doi:10.2217/epi-2017-0055
21. Liang HF, Zhang XZ, Liu BG, Jia, G-T., Li, W-L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566–1576.
22. Lu L, Sun J, Shi P, Kong W, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8:44096–44107. doi:10.18632/oncotarget.17307
23. Tang YY, Zhao P, Zou TN, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36:901–908. doi:10.1089/dna.2017.3862
24. Yang E, Du WW, Li X, Yee, A J., Yang, B B. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–3931. doi:10.1038/onc.2015.460
25. Kong Z, Wan X, Zhang Y, et al Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;493:1217–1223. doi:10.1016/j.bbrc.2017.07.162
26. Amant F, Mirza MR, Koskas M, Creutzberg, C L. Cancer of the corpus uteri. Int J Gynaecol Obstet. 2018;143 Suppl 2:37–50. doi:10.1002/ijgo.12612
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.