Back to Journals » Infection and Drug Resistance » Volume 15
Analysis of Metagenomic Next-Generation Sequencing Results of 25 Pus Samples
Authors Shi Y, Wu J , Liu T, Yue L, Liu Y, Gu Y, Qi Y
Received 11 August 2022
Accepted for publication 27 October 2022
Published 8 November 2022 Volume 2022:15 Pages 6515—6524
DOI https://doi.org/10.2147/IDR.S385925
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yuru Shi, Jing Wu, Ting Liu, Li Yue, Yang Liu, Yan Gu, Yingjie Qi
Department of Clinical Laboratory, Infection Hospital Area of the First Affiliated Hospital of University of Science and Technology of China (Hefei Infectious Disease Hospital), Hefei, 230022, People’s Republic of China
Correspondence: Yingjie Qi, Department of Clinical Laboratory, Infection hospital area of the First Affiliated Hospital of University of science and technology of China (Hefei infectious disease hospital), No. 218 Susong Road, Hefei, Anhui Province, 230022, People’s Republic of China, Email [email protected]
Purpose: To explore the clinical value of detecting pathogens in pus samples by metagenomic next-generation sequencing (mNGS).
Methods: The 25 pus samples from infected patients were collected in this research. The positive rate and consistency of pathogenic bacteria detected by mNGS and conventional methods were compared. The pathogen types detected by the two methods were analyzed. Furthermore, the modifications of antibiotic treatment therapy were also evaluated based on mNGS results.
Results: The sensitivity of mNGS method in detecting pathogenic bacteria in pus samples was better than that of conventional method (96% vs 40%; P < 0.01). Only 10 samples were detected pathogens by conventional methods, but 24 samples were detected by mNGS method. In specific, the results of conventional methods showed 10 samples had 11 kinds of pathogenic bacteria, of which 9 samples were single pathogen and 1 sample had two kinds of pathogenic bacteria. The results of mNGS method showed 24 samples were detected with 54 kinds of pathogenic bacteria, of which 15 samples were detected with single pathogen, and 9 samples were detected with two or more kinds of pathogenic bacteria. The two methods had 9(36%) consistent results, 14 (56%) completely different results, and 2 (8%) partially consistent results, and the kappa value was 0.19. Notably, mNGS could detect viruses, anaerobic bacteria, and other uncommon pathogens simultaneously.
Conclusion: The application of mNGS in the detection of pus specimens from different parts not only have high accuracy rate and also reduce the turnaround time of diagnosis. In addition, the performance of mNGS detection of anaerobic bacteria and caustic bacteria is better than conventional methods. The mNGS diagnosis in pus sample may play an important role in clinical diagnosis and treatment strategy decisions.
Keywords: mNGS, pus specimens, pathogen
Introduction
In most cases, the formation of pus is associated with an infection that can involve the skin, liver, lungs, brain, eyes, and joint cavities. Once infection occurs, clinicians are prone to choose antibiotics as anti-infective treatments. Rapid and accurate detection of pathogens is particularly important for patient treatment and could reduce antibiotic abuse.1 Conventional methods are less accurate and time-consuming, which may result in inadequate treatment, especially for anaerobic bacteria and difficult-cultured pathogens tests. mNGS technology is a revolutionary diagnostic technology, which could comprehensively detect pathogens without conventional.2 Nucleic acids are extracted directly from specimens for unbiased detection.3 In this way, pathogen sequences with low abundance can also be obtained from specimens with high sensitivity.4
In the clinic, mNGS was first used to detect leptospirosis in cerebrospinal fluid samples of a 14-year-old boy with severe immunodeficiency and continuous fever for 4 months.5 Since then, the application of metagenomic sequencing technology in the detection of infectious diseases has been continuously reported.6,7 However, the clinical applications of mNGS in pus samples from infected patients are rarely reported and clinical value is unclear.8 In this study, mNGS was applied to pus samples to evaluate the sensitivity and clinical benefits of this method, and compared with conventional methods.
Materials and Methods
General Information
Twenty-five patients from the First Affiliated Hospital of the University of Science and Technology of China were enrolled from April 2020 to November 2021 as research objects. The inclusion criteria were as follows: (1) The patients or authorized family member agrees to carry out mNGS testing; (2) ≥18 years old; (3) The clinician suspected the case of infection, and the corresponding antibiotic treatment is effective. Exclusion criteria: (1) Pregnant and the patients under the age of 18; (2) Samples collected were insufficient for conventional and mNGS tests; (3) Non-infectious pus specimens. Specimens from the same patient were used by conventional methods and mNGS simultaneously. This study was a retrospective study, and patient data were anonymously numbered. The study does not involve patient privacy and conforms to the principles of medical ethics. Demographic information, basic diseases, symptoms of infection, and antibiotic treatment were recorded.
Research Methods
Collection of Patient Samples
Standardized aseptically collected specimens from abscess sites of 25 patients were placed into nucleic acid-free containers. The samples were tested by conventional methods (culture and smear), and mNGS was tested at the same time. Put specimens into the nucleic acid-free container and were stored at −80°C before testing. The patient’s medical records and test results were analyzed.
DNA Extraction
Except plasma samples, the other samples were prepared for wall-breaking reaction, and 200 ng DNA was extracted using the TIANamp Micro DNA Kit (DP316, Tiangen Biotech).
Construction of DNA Libraries and Sequencing
DNA libraries were constructed through DNA-fragmentation, end-repair, adapter-ligation and PCR amplification. Qualified libraries were pooled; DNA Nanoball (DNB) was fabricated from single-stranded DNA circles, and sequenced by MGISEQ-2000 platform.
Bioinformatic Analysis
High-quality sequencing data were generated by removing low-quality reads, and short (length <35bp) reads at first, followed by computational subtraction of human host sequences mapped to the human reference genome (ftp://ftp.ncbi.nlm.nih.gov/genome). The remaining sequence data were aligned to microbial genome databases by BWA (Burrows-Wheeler alignment, http://bio-bwa.sourceforge.net). The database used for the present study contains 2328 bacterial species, 199 fungal species, 4189 viruses, 135 parasites, 83 mycobacterium, and 41 mycoplasma/chlamydia, they were associated with human diseases. Criteria for a positive mNGS result: (1) Bacteria, fungi, mycoplasma, chlamydia, Rickettsia G_SMRN or SDG_SMRN ≥3, species level Latin_SMRN or SDLatin_SMRN ≥3; (2) Mycobacterium tuberculosis complex: genus level G_SMRN or SDG_SMRN ≥1, species level Latin_SMRN or SDLatin_SMRN ≥1; (3) Virus: species level G_SMRN or SDG_SMRN ≥3, type or subtype Latin_SMRN or SDLatin_SMRN ≥1; (4) Parasites: genus level G_SMRN or SDG_SMRN 100, species level Latin_SMRN or SDLatin_SMRN ≥ 100; (5) Fungi and parasites were filtered according to DepthDepth_Ratio ≥0.5 and Shannon_index ≥0.75.
mNGS Positive Standard
The following criteria were used to identify favorable outcomes because to the lack of guidelines for interpreting mNGS findings and the wide range of various sequencing platforms: (1) having pathogenicity from pus sample that has been noted in the literature; (2) Bacteria: >30% relative abundance for opportunistic pathogenic bacteria at the genus level, 1 stringently mapped read for Mycobacterium TB complex, and non-tuberculous mycobacteria. (3) fungus: 1 stringently mapped read at the species level; 10 stringently mapped reads for mold at the species level. (4) 1 stringently mapped read at the species level for mycoplasma and chlamydia.
Statistical Analysis
SPSS 23.0 statistical software was used to perform the data analysis, the counting data is expressed by the number of cases and rates, and the measurement data is expressed by mean ± standard deviation. Comparative analysis was conducted by Fisher’s precision probability test and Kappa test, p-values of <0.05 were considered statistically significant difference. If the Kappa value is ≧ 0.75, the two detection systems have good equivalence; if <0.4, the detection system does not have equivalence.
Results
Basic Situation Analysis of Patients
The basic clinical characteristics of the patients are shown in Table 1. Among the 25 patients, there were 16 males and 9 females, with an average age of 54.36±16.58 years, and the majority of them were 30–60 years old (64%). Eighteen patients had basic diseases (80%), including acquired immune deficiency syndrome, malignant tumor, diabetes, hypertension and 6 patients underwent surgery. A total of 25 specimens were collected, including 6 whole blood specimens, 1 skin tissue specimen, 1 drained pus specimen, and 15 wound secretion swab specimens. Clinical symptoms were manifested as pain (13 cases, 44.83%), redness and swelling (7 cases, 24.14%), as well as fever (4 cases, 13.79%), cough (3 cases, 10.34%) and fester (2 cases, 6.90%). Twenty patients (80%) improved significantly after treatment, 2 patients (8%) were stable, and 3 patients (12%) had no improvement (Table 1).
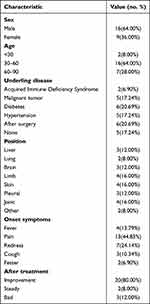 |
Table 1 Patient Characteristic Analysis |
Analysis of mNGS and Conventional Methods
In 25 specimens, mNGS test results showed that 1 case was negative and 24 cases were positive. The test results of using conventional methods showed that 15 cases were negative, 10 cases were positive. The positive rate of mNGS was higher than that of conventional methods (96% vs 40%, p < 0.01) (Table 2). Different types of pathogens detected by mNGS and conventional methods are shown in Figure 1. Forty-eight kinds of bacteria were detected through mNGS, among which 7 strains of streptococcus and 5 strains of Clostridium ranked first and second, respectively. The conventional methods detected 10 strains of bacteria, the most common pathogens were 2 strains of streptococcus and 1 strain of fungus; however, there was no virus was detected using conventional methods, only detected by the mNGS. mNGS identified several uncommon pathogens in patients with SSTI, such as Nocardia Pythium, Vibrio vulnificus, and Treponema pallidum. The concordance rate between mNGS and conventional methods in the identification of pathogens in pus samples were 9 cases (36%). Two cases (8%) were not completely consistent between the two test methods, and 14 cases (56%) were completely inconsistent, and the kappa value was 0.19 (Figure 1, Table 3). As shown in Figure 2, mNGS detected a total of 54 strains of pathogens, including 48 strains of bacteria (26 strains of anaerobic bacteria and 22 strains of aerobic bacteria), 1 strain of spirochete, 4 strains of fungi and 1 strain of virus (Figure 3). Multiple pathogens were detected in 4.16% of cases by culture, while mNGS showed a significantly higher proportion of multiple microbial infections (37.5%, P = 0.004) (Figure 4). Among the cases of multi pathogen infection, mNGS detected 9 cases of mixed infection of anaerobic bacteria, 1 case of virus infection (Varicella zoster virus) mixed with bacteria.
 |
Table 2 Comparison of Positive Rate Between Two Methods |
 |
Table 3 Comparison of mNGS Results with Traditional Culture Results |
 |
Figure 1 Comparison of the mNGS results with the traditional detection methods. |
 |
Figure 2 Detection Results of the mNGS. |
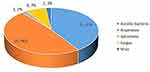 |
Figure 3 Pathogen composition detected by mNGS. |
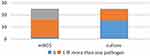 |
Figure 4 Proportion of multiple pathogens, detected by mNGS and culture. |
Conventional Method Was Negative, and the Use of Antibiotics After the Use of mNGS and Its Outcome
Among the 15 samples with negative results of conventional methods, 14 samples were detected with mNGS, of which P7 was not adjusted for antibiotics after discharge due to not waiting for the report results. Thirteen cases were treated with antibiotics empirically, and targeted treatment was carried out according to the detection results of mNGS, and the infection of 12 patients was controlled (Table 4).
 |
Table 4 The Traditional General Culture Was Negative, and the Use of Antibiotics After the Use of mNGS and Its Outcome |
Analysis of Typical Cases
Patient No. 22 developed symptoms of difficulty in mouth opening, inability to swallow food, involuntary extension of head and neck, stiffness of the limbs, and paroxysmal convulsions after admission. Physical examination found that the right lower limb had chronic skin ulcers. The patient broke repeatedly during 30 years. We empirical use of ceftriaxone sodium for antiinfection treatment. Cerebrospinal fluid, plasma and lower extremity ulcerated pus of the patients were collected to perform mNGS sequencing analysis and conventional methods. After 20 hours, the result of mNGS displayed, no microorganism in the cerebrospinal fluid and plasma, nevertheless, Bacteroides pyogenes, Porphyromonas soxerii, Morganella morganii, Clostridium nucleatus, Streptococcus anaerobic digesticosis, and Clostridium tetanus were found in the pus sample by mNGS (Figure 5). After identifying the pathogen infection, we immediately adjust the treatment plan and use anaerobic anti-infection targeted therapy-metronidazole and penicillin, actively carry out debridement. After treatment, the patient’s fever time was shortened, but there was still intermittent fever. After 48 h, the results of conventional methods show that only detected staphylococcus hominis in the pus of lower limb ulcer. Because the patient was sent to hospital late for treatment, respiratory failure, convulsions, poor nutritional status and other complications occurred, and the patient’s family members gave up treatment.
 |
Figure 5 Patient 22 all pathogens detected in pus samples. |
Discussion
The rapid diagnosis of pus pathogens in infected sites has always been an urgent problem in clinical practice.9 mNGS has been used to detect pathogens in a variety of infectious diseases, including encephalitis,10 prosthetic joint infection11 and infection in immunocompromised patients.12,13 However, the detection of pathogens by mNGS in pus specimens from infected patients has not been well studied. In this study, the clinical diagnostic performance of mNGS in pus samples was evaluated concurrently with conventional methods.
The clinical symptoms of the patients include local and systemic infection. Patients with diseases accounted for 84% of the pus specimens collected, such as primary lesion infection, immune deficiency, tumor and other diseases, reflecting the complexity of clinical patients’ conditions. If the infection is not diagnosed and treated promptly under such circumstances, it can lead to a poor prognosis and even death.14 Although the smear method can quickly know whether there is a pathogen detected, it is not clear what the pathogen is. Conventional method takes a long time, and ordinary bacterial culture takes up 24 hours at the earliest, and fungal culture takes up to 5 days. If it is identification of mixed pathogen infection, it needs to be purified first, and the culture time is longer. The smear method is fast, but does not accurately identify the pathogen. Currently, the mNGS method can generate report within 24 hours to achieve the purpose of rapid detection and treatment.15 Furthermore, 80% of patients have improved or stabilized infection after symptomatic treatment with antibiotics. It is suggested that timely symptomatic anti-infective treatment can minimize the occurrence of adverse prognostic events.
In this research, we compared the two detection methods of mNGS and conventional methods, and the results indicated that (1) the positive rate of mNGS is significantly higher than conventional. mNGS has high sensitivity and the detection process is not affected by the use of antibiotics.16 For anaerobic and caustic bacteria that are difficult to cultivate, the detection rate of mNGS is higher than that of conventional methods.17,18 (2) mNGS identified more pathogens than conventional, especially detected anaerobes, viruses and other uncommon pathogens. On the basis of completely inconsistent results, no pathogen was detected in 15 cases using conventional methods, while caustic bacteria and anaerobic bacteria were detected by mNGS. According to the composition of pathogens detected by mNGS in pus samples, the detection rate of anaerobic bacteria, spirochete and virus is 52%, which is consistent with the research results mNGS has advantages in rapid diagnosis of caustic bacteria, anaerobic bacteria and time-consuming bacteria.19 In addition, mNGS showed better ability to recognize co-pathogens in a single specimen than conventional, which is consistent with other results.20,21
Anaerobic bacteria (48%) were most frequently detected at the infected site of patients, followed by aerobic bacteria (41%), as well as fungi, viruses and spirochetes. According to the 2014–2019 bacterial drug resistance monitoring report of clinical wound and pus samples from the National Bacterial Drug Resistance Monitoring Network, the main pathogens in wound and pus samples are Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae and Pseudomonas aeruginosa.22 The conventional method for pathogen identification are affected by many factors such as environment, medium, and time. The ability to detect pathogens is limited because pathogens could only be successfully cultured and identified under appropriate conditions. However, in clinical practice, the detection of anaerobic bacteria is difficult to be implemented in the conventional microbiology laboratory due to the strict anaerobic environment for specimen collection and transportation. Moreover, virus testing is not routine in clinical practice. mNGS extracts all nucleic acids from the abscess site samples of patients and compares them with nucleic acid sequences from gene libraries to determine the type of pathogenic microorganisms. It does not depend on the conventional environment and is not affected by antibiotic use, making it an unbiased test.16
Some scholars proposed that the pathogens in pus specimens may be the cause of infectious diseases, but the high background of human genomes in pus specimens may reduce the sensitivity and specificity of sequencing.23 The typical case No. 22 showed that the pathogen load of local lesions was higher than cerebrospinal fluid, plasma and pus samples. The detection of Clostridium tetanus, an important infectious pathogen in pus specimens of local infection, provides an important experimental basis for clinical practice. At the same time, some research used Illumina and Nanopore to sequence 87 body fluid samples from 77 patients with acute infection, and compared them with the clinical adjudication, culture, and PCR testing. The results showed that the sensitivity and specificity of mNGS for bacterial detection were 81–86% and 91–95%, and for fungal detection were 63–70% and 92–96%, respectively.24 In this study, mNGS was performed for 7 patients who had paired body fluid and plasma samples, and a total of 9 pathogens were detected, and it was found that cfDNA burden of the pathogen in local body fluid was 160-fold higher than that in plasma. Eight pathogens (88.9%) were detected in body fluids, while only three pathogens (33.3%) were detected in plasma. These results indicate that the detection of pathogenic microorganisms is more sensitive in locally infected specimens than in plasma specimens, and therefore, pus specimens do not limit the detection of pathogenic microorganisms due to the high background of the human genome.
It is worth noting that 14 specimens were diagnosed positive only by mNGS, but negative by conventional.13 patients were treated with previous antibiotic experience, changed from the previous antibiotic full coverage treatment to targeted treatment for pathogens, and 12 patients showed improvement in clinical infection symptoms during the 30 days follow-up. Adjust the treatment after antibiotics, reduce the use of antibiotics, achieve the purpose of precise use of antibiotics, and timely and accurate antibiotic treatment has an important impact on the prognosis of patients.25
This study has some limitations. Due to the limitation of the number of specimens, the statistical results may be biased, and the number of specimens will be increased in the future. At the same time, RNA sequencing or drug resistance data were not examined, so the detection of RNA viruses may be affected.
To sum up, we have reported the performance of mNGS on identifying pathogens in pus specimens. As a new detection method, mNGS not only has the characteristics of high sensitivity, high accuracy and fast speed, but also has a good ability to detect the pathogens of mixed infection, which has more advantages than conventional methods.
Data Sharing Statement
All data and materials were in full compliance with the journal’s policy.
Ethics Approval and Informed Consent
This study was completely approved by the institutional review board of the First Affiliated Hospital of University of Science and Technology of China (IRB number: 2021-BE(H)-005). This study is a retrospective study and we could not obtain written informed consent from the patients who have been discharged from hospital. Therefore, we obtained verbal informed consent from all participants during the telephone follow-up with the approval of the institutional review board of the First Affiliated Hospital of University of science and technology of China. All data that could indicate the identity of the patients were kept strictly confidential in this study. Work on human beings is conducted in accordance with the Declaration of Helsinki.
Consent for Publication
We have obtained consent to publish from all the participants.
Acknowledgments
Thanks to BGI technicians for their guidance on this article.
Funding
This study was supported by Single Center Study on Respiratory Pathogen Detection (fund project no. HXD202103).
Disclosure
All authors declare that they have no conflicts of interest in this work.
References
1. Zhao Y, Chen J, Bai B, et al. Pathogen determination from clinical abscess fluids using metagenomic next-generation sequencing. Folia Microbiol. 2021,66(2):197–202. doi: 10.1007/s12223-020-00829-x.
2. Wang Q, Miao Q, Pan J, et al. The clinical value of metagenomic next-generation sequencing in the microbiological diagnosis of skin and soft tissue infections. Int J Infect Dis. 2020;100:414–420 doi: 10.1016/j.ijid.2020.09.007.
3. Han D, Li Z, Li R, et al. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol, 2019,45(5–6):668–685. doi: 10.1080/1040841X.2019.1681933.
4. Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. doi:10.1038/s41564-018-0349-6
5. Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370(25):2408–2417. doi:10.1056/NEJMoa1401268
6. Tang W, Zhang Y, Luo C, et al. Clinical application of metagenomic next-generation sequencing for suspected infections in patients with primary immunodeficiency disease. Front Immunol. 2021;12:696403. doi:10.3389/fimmu.2021.696403
7. Liu W, Fan Z, Zhang Y, et al. Metagenomic next-generation sequencing for identifying pathogens in central nervous system complications after allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2021;56(8):1978–1983. doi:10.1038/s41409-021-01243-8
8. Guo LY, Feng WY, Guo X, et al. The advantages of next-generation sequencing technology in the detection of different sources of abscess. J Infect. 2019;78(1):75–86. doi:10.1016/j.jinf.2018.08.002
9. Venkatesan A. Encephalitis and brain abscess. Continuum. 2021;27(4):855–886. doi:10.1212/CON.0000000000001006
10. Brown JR, Bharucha T, Breuer J. Encephalitis diagnosis using metagenomics: application of next generation sequencing for undiagnosed cases. J Infect. 2018;76(3):225–240. doi:10.1016/j.jinf.2017.12.014
11. Cai Y, Fang X, Chen Y, et al. Metagenomic next generation sequencing improves diagnosis of prosthetic joint infection by detecting the presence of bacteria in periprosthetic tissues. Int J Infect Dis. 2020;96:573–578. doi:10.1016/j.ijid.2020.05.125
12. Parize P, Muth E, Richaud C, et al. Untargeted next-generation sequencing-based first-line diagnosis of infection in immunocompromised adults: a multicentre, blinded, prospective study. Clin Microbiol Infect. 2017;23(8):574.e1–574.e6. doi:10.1016/j.cmi.2017.02.006
13. Peng JM, Du B, Qin HY, Wang Q, Shi Y. Metagenomic next-generation sequencing for the diagnosis of suspected pneumonia in immunocompromised patients. J Infect. 2021;82(4):22–27. doi:10.1016/j.jinf.2021.01.029
14. Xie G, Zhao B, Wang X, et al. Exploring the clinical utility of metagenomic next-generation sequencing in the diagnosis of pulmonary infection. Infect Dis Ther. 2021;10(3):1419–1435. doi:10.1007/s40121-021-00476-w
15. Zhang HC, Ai JW, Cui P, et al. Incremental value of metagenomic next generation sequencing for the diagnosis of suspected focal infection in adults. J Infect. 2019;79(5):419–425. doi:10.1016/j.jinf.2019.08.012
16. Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(suppl_2):S231–231S240. doi:10.1093/cid/ciy693
17. Zhao Y, Chen J, Bai B, et al. Pathogen determination from clinical abscess fluids using metagenomic next-generation sequencing. Folia Microbiol (Praha). 2021;66(2):197–202. doi:10.1007/s12223-020-00829-x
18. Zhou H, Larkin P, Zhao D, et al. Clinical impact of metagenomic next-generation sequencing of bronchoalveolar lavage in the diagnosis and management of pneumonia: a multicenter prospective observational study. J Mol Diagn. 2021;23(10):1259–1268. doi:10.1016/j.jmoldx.2021.06.007
19. Li H, Gao H, Meng H, et al. Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front Cell Infect Microbiol. 2018;8:205. doi:10.3389/fcimb.2018.00205
20. Wang J, Han Y, Feng J. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med. 2019;19(1):252. doi:10.1186/s12890-019-1022-4
21. Fang X, Mei Q, Fan X, et al. Diagnostic value of metagenomic next-generation sequencing for the detection of pathogens in bronchoalveolar lavage fluid in ventilator-associated pneumonia patients. Front Microbiol. 2020;11:599756. doi:10.3389/fmicb.2020.599756
22. China Antimicrobial Resistance Surveillance System. Antimicrobial resistance of bacteria from wound and pus specimens: surveillance report from China Antimicrobial Resistance Surveillance System in 2014-2019. Chin J Infect Control. 2021;20(02):145–156.
23. Miller S, Naccache SN, Samayoa E, et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29(5):831–842. doi:10.1101/gr.238170.118
24. Gu W, Deng X, Lee M, et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med. 2021;27(1):115–124. doi:10.1038/s41591-020-1105-z
25. Wang Q, Miao Q, Pan J, et al. The clinical value of metagenomic next-generation sequencing in the microbiological diagnosis of skin and soft tissue infections. Int J Infect Dis. 2020;100:414–420. doi:10.1016/j.ijid.2020.09.007
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
