Back to Journals » International Journal of General Medicine » Volume 15
Analysis of Key Genes for Slow Transit Constipation Based on RNA Sequencing
Authors Yu L , Yang X, Guan W, Zhang D, Ren S, Xing Y, An D, Zhang J, Zhu Y, Zhu A
Received 6 July 2022
Accepted for publication 16 September 2022
Published 28 September 2022 Volume 2022:15 Pages 7569—7579
DOI https://doi.org/10.2147/IJGM.S380208
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Linfeng Yu1 *, Xiuding Yang1 *, Wenlong Guan,1 Dongxu Zhang,1 Shuo Ren,2 Yanwei Xing,1 Da An,3 Jian Zhang,1 Yuekun Zhu,1 Anlong Zhu1
1Department of General Surgery, The First Affiliated Hospital of Harbin Medical University, Harbin, People’s Republic of China; 2Department of Gastrointestinal Surgery, Sichuan Cancer Hospital, Chengdu, People’s Republic of China; 3Department of Anesthesiology, The First Affiliated Hospital of Harbin Medical University, Harbin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Anlong Zhu, Department of General Surgery, The First Affiliated Hospital of Harbin Medical University, Harbin, People’s Republic of China, Tel/Fax +86 13504848555, Email [email protected]
Purpose: This study aims to identify key genes in slow transit constipation (STC). We also sought to explore the potential link between STC and colorectal cancer.
Patients and Methods: mRNA expression profiles were obtained by RNA sequencing, and differentially expressed genes were identified. Functional enrichment analysis and a protein–protein interaction (PPI) network was explored, and differentially expressed genes common to STC and colorectal cancer were examined. Analysis of the effect of constipation and colorectal cancer common genes on the overall survival of colorectal cancer patients based on GEPIA database.
Results: Functional enrichment showed that significantly different genes are related to lymphocyte chemotaxis, positive regulation of inflammatory response, cellular response to tumor necrosis factor, extracellular region, extracellular space and chemokine activity. The hub gene for STC was found in the PPI network. In addition, AQP8 and CFD were common differential genes for STC and colorectal cancer. AQP8 affects overall survival in patients with colorectal cancer.
Conclusion: Our findings will contribute to understanding the pathology of STC at the molecular level, with the first discovery that AQP8 may be a hub gene in the transition from STC to colorectal cancer.
Keywords: slow transit constipation, RNA sequencing, hub genes, colorectal cancer, AQP8
Introduction
Constipation is a common gastrointestinal disease, with an incidence rate of 14% worldwide and 10–15% in China.1,2 Constipation is more common in females, and its incidence increases with age.3 As a functional gastrointestinal disease, constipation is characterized by infrequent bowel movements, defecation difficulty, and incomplete bowel evacuation sensation.4 It has been reported that constipation is associated with inflammatory bowel disease, irritable bowel syndrome, and relevant comorbidities.5 Among them, the most common is slow transit constipation (STC) caused by colonic transit dysfunction.6
Overall, the pathogenesis of STC remains elusive. Unhealthy living habits and genetic factors promote the occurrence of STC.7 Some studies have proposed that STC may be related to Cajal interstitial cells, enteric nervous system dysfunction, and abnormal hormone levels.8–10 STC is attracting increasing attention from medical researchers due to its serious impact on the physical and mental health and quality of life of patients.11
Constipation has been proposed as one potential risk factor for colorectal neoplasia. A biologic plausibility for this link is thought to be an increase in contact time between stool carcinogens and colonic mucosa. Although many studies have examined the relationship between colorectal neoplasia and constipation, no consensus has been reached. In a long-term follow-up study of 2165 patients with constipation, Eun Mi Song et al found that only 5 people were newly diagnosed with colorectal cancer during more than 10 years of follow-up, with and without slow transit constipation. Colorectal cancer risk was not significantly different (P=0.575).12 In a study in the Netherlands, frequent bowel movements were associated with an increased risk of rectal cancer in men, and constipation was associated with a decreased risk.13 The discrepancy in these results is likely due to the multiple inherent limitations that exist when evaluating the complex potential relationship of constipation and CRC. Jessica Citronberg et al found in a prospective study that long-term use of nonfibrotic laxatives in patients with STC would increase the risk of colorectal cancer.14 It has also been reported that STC does not directly lead to the occurrence of colorectal cancer but that it increases the incidence of colorectal polyps, which may become cancerous.
There is to date no consensus regarding the treatment of STC, and individualized treatment is generally carried out.15 The first step is to change lifestyle, medication, and psychotherapy. Patients treated conservatively usually take long-term oral laxatives. If conservative treatments for STC are ineffective, surgical treatment may be necessary.16,17 Many young patients undergo subtotal resection or total resection of the colon. Therefore, it is urgent to explore the pathological mechanism of STC. The purpose of our study was to identify hub genes that affect the pathogenesis of STC through bioinformatics methods based on RNA sequencing. We identified the hub gene that affects STC and attempted to reveal the correlation between STC and colorectal cancer.18 In addition, we explored the effect of constipation and colorectal cancer common genes on the prognosis of colorectal cancer patients.
Materials and Methods
Basic Patient Information
We strictly follow the Rome IV standard for research subject inclusion.19 All patients in the case group were confirmed to have STC by imaging prior to surgery. The postoperative histopathological evaluation standard was a reduction in interstitial cells of Cajal. The exclusion criteria were as follows: (1) chronic constipation caused by spastic pelvic floor syndrome, rectocele, megacolon, or other organ lesions; (2) obvious gastric intestinal transit dysfunction; (3) secondary constipation caused by drugs, endocrine, metabolic, or neurological diseases, renal failure, cirrhosis, severe hypertension, and severe obesity or a body mass index exceeding 30; and (4) incomplete data that influenced the diagnosis. Only patients who were diagnosed with slow colonic transit function and did not meet the exclusion criteria were included in this study. Finally, the diseased colons of 3 patients with STC were used as the experimental group, and paracancerous tissues were used as the control group (Table 1).
 |
Table 1 Basic Clinical Information of STC and Control Group Patients |
This study was approved in writing by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University, and all participants signed an informed consent form. The acquisition of STC tissues and normal adjacent tissues complies with the Declaration of Helsinki.
RNA Isolation and Sequencing
Total RNA was isolated from samples by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). A NanoPhotometer spectrophotometer (IMPLEN, CA, USA) was used to assess sample purity. A Qubit 3.0 A fluorometer (Life Technologies, CA, USA) was employed to detect the RNA concentration and an Agilent 2100 RNA Nano 6000 Assay Kit (Agilent Technologies, CA, USA) to detect RNA integrity. Messenger RNA (mRNA) was oligo-d (T) purified for polyA selection. The purified mRNA was fragmented in fragmentation buffer, and complementary DNA (cDNA) was generated. End repair and adapter ligation were performed after purification using the QIAquick PCR purification kit. Agarose gel electrophoresis was performed to recover target fragments, after which PCR amplification was performed to complete the library preparation. Next, the effective concentration of the library was accurately quantified to ensure its quality. Finally, we used the Illumina platform for sequencing with the PE150 strategy.
Sequencing Data Quality Control
Sequencing was performed on the Illumina Nova 6000 platform with 150bp double end reads to obtain a minimum yield of 20M reads per sample. Sequence data were processed using hisat2 and mapped to the human reference genome (Homo_sapiens.GRCh38.94.chr).
Differentially Expressed Genes in STC
In RNA-Seq analysis, the expression level of genes is evaluated by counting sequencing sequences (reads) located in the genomic region or exon region. Fragments per kilobase per million mapped (FPKM) is a very effective tool for quantitatively estimating gene expression using RNA-Seq technology, whereby the gene expression value is based on the number of fragments from a gene per kilobase length per million fragments. Differential gene screening mainly refers to the multiple of difference (fold change value) and P value as related indicators. Differential expression analysis was performed using the DESeq2 R package. In this study, the threshold for a significant difference in gene expression was set as an absolute value of log2-fold change > 1 and a P value < 0.05. A heatmap of significantly different genes between the STC group and the control group was produced using the “pheatmap” package of R software (version: x64 4.1.1). Significant upregulation and downregulation of differentially expressed genes are illustrated as a volcano map.
Differentially Expressed mRNA Functional Enrichment in STC
We selected mRNAs among all significantly differentially expressed genes. To study the biological functions of these differentially expressed mRNAs, we used the online tool Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov/) to assess enrichment as GO and KEGG functions, and we generated a bubble chart with Sangerbox (http://sangerbox.com/Tool). For GO term enrichment, we used FDR<0.05 as the significance criterion. We chose the top ten mRNAs with the smallest FDR value for the bubble chart. For KEGG pathway enrichment, we used q<0.05 as the significant criterion, and we selected the top ten with the smallest q value for bubble chart preparation.
Protein–Protein Interaction Network in STC
We filtered out noncoding RNAs and transcription factors from the significantly differentially expressed genes. To study interrelationships between meaningful proteins, we used the online database STRING (https://string-db.org/) to construct a protein–protein interaction network; certain key genes in the protein–protein interaction network was also identified. We used the CytoHubba plug-in to screen hub genes. The relationship between genes is expressed by the degree of connection, which is also the standard for identifying hub genes. We chose the top ten genes with the highest correlation as hub genes. Cytoscape software (version: 3.6.0) was applied to visualize the results of the protein–protein interaction network.
Correlation Between STC and Colorectal Cancer
To explore whether there is a correlation between STC and colorectal cancer, gene expression microarray data related to colorectal cancer were downloaded from the Gene Expression Omnibus (GEO) database. Gene expression microarray chips were used to screen genes differentially expressed between colorectal cancer and adjacent normal tissues. We obtained significantly different genes for colorectal cancer in the GSE146587 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146587) and GSE113513 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113513) datasets and set LogFC (fold change) >2 and adj P value <0.05 as the standard screening criteria for significantly different genes. The differential genes screened in the two gene sets and those screened in the STC patients were crossed-referenced to obtain common genes. A Venn diagram showing the overlap between genes differentially expressed in colorectal tumors and STC was generated using FunRich software (version: 3.1.1).
GEPIA Database to Verify the Impact of Common Genes on the Prognosis of Colorectal Cancer Patients
We used the Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) database to analyze the effect of the screened constipation and colorectal cancer common genes on the prognosis of colorectal cancer patients. On the main page, we select “Single Gene Analysis”, then enter the gene to be analyzed, and click “Survival Analysis”. Select the colorectal cancer to be analyzed at “Datasets Selection”, Group Cutoff defaults to “Median”, Cutoff-High(%) and Cutoff-Low(%) default to 50, click “Plot” to generate a survival curve graph. Statistical significance was set at P<0.05.
Results
Differentially Expressed mRNA in STC
We used the fold change method to screen for differential genes. By setting the conditions |log2-fold change|≥1 and P<0.05, we obtained 509 differentially expressed mRNAs (329 upregulated and 180 downregulated). We produced a heatmap of all differentially expressed mRNAs in R language. (Figure 1) A volcano map was used to illustrate upregulation and downregulation of differential genes. (Figure 2)
Differentially Expressed mRNA Functional Enrichment in STC
DEGs were divided into the three functional categories of cell component, molecular function, and biological process, and genes were enriched in functional terms as much as possible. Enrichment analysis will provide a P value for the term involved according to the hypergeometric distribution relationship: the smaller the P value, the more significant the enrichment of differential genes in this term. According to GO enrichment, lymphocyte chemotaxis, positive regulation of inflammatory response, cellular response to tumor necrosis factor, extracellular region, extracellular space, high-density lipoprotein particle, chemokine activity and retinol binding were the most significant GO terms. Neuroactive ligand–receptor interaction, cytokine–cytokine receptor interaction and rheumatoid arthritis were the most meaningful KEGG pathways. The enrichment results of GO and KEGG are shown in Figure 3.
Protein–Protein Interaction Network (PPI) in STC
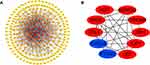
In total, 246 upregulated mRNAs and 108 downregulated mRNAs were identified in this study. To explore the relationship between all differential mRNAs, we used Cytoscape software to determine the PPI of the mRNAs. In the obtained network, the number of nodes was 349, the number of edges was 835, and the average node degree was 4.79. The top 10 nodes with the highest degree of PPI network connectivity were identified as hub genes using the CytoHubba plugin in Cytoscape: CXCL8, LEP, TYROBP, PTGS2, AGT, APOE, CCL4, SPI1, IGF1 and ADIPOQ. (Figure 4, Table 2)
 |
Table 2 Ten Hub Genes Identified by CytoHubba |
Relationship Between STC and Colorectal Cancer
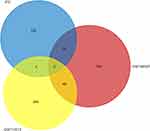
Through the Venn diagram, we intersect the significantly differentially expressed genes of colorectal cancer obtained from the GSE146587 and GSE113513 data sets with the differential genes in STC. There are only two genes in the intersection of the three data sets, AQP8 and CFD. FunRich 3.1.3 software was used to draw the Venn diagram shown in Figure 5.
Influence of Constipation and Colorectal Cancer Common Genes on Prognosis of Colorectal Cancer Patients
In the GEPIA database, we analyzed the prognostic impact of constipation and colorectal cancer common genes on colorectal cancer patients. The results showed that the gene AQP8 (P=0.02) had an effect on the overall survival of colorectal cancer patients. In addition, gene CFD (P=0.92) had no effect on overall survival in patients with colorectal cancer (Figure 6).
Discussion
STC is the major cause of chronic constipation. Although it does not directly affect the life of patients, it greatly reduces quality of life. The disease imposes a serious psychological burden on the patient, which can lead to depression. In this study, we found genes abnormally expressed in STC tissues to explore its pathogenesis at the molecular level.
GO analysis involves three aspects: molecular function, biological process, and cell component. Differential genes are mainly involved in “biological regulation”, “regulation of cellular process”, “plasma membrane”, “vesicle”, “extracellular vesicle”, “receptor binding” and “receptor activity” terms. STC is a multifactorial disorder with a complex pathogenesis. Amelia Mazzone et al found that miRNA targeting SCN5A resulted in decreased smooth muscle motility.20 And a study by Wang et al showed that electrical stimulation of the colon can be used as an effective treatment for STC. Indeed, colonic electrical stimulation can promote vesicle production by intramuscular nerves and colon movement. We infer that vesicles play a role in STC.21 Abnormal signal transduction is also involved in the pathogenesis. Progesterone interacts with progesterone receptor B (PGR-B), a G protein-coupled receptor, to impair muscle contraction.22 KEGG analysis revealed enrichment of the NF−kappa B signaling pathway, which is consistent with the conclusion of Zhan et al, who conducted a study on the mechanism of action of Maren pills in the treatment of constipation. Maren pills promote expression of AQP3 by inhibiting the NF-κB pathway.23
To explore interaction between the detected differentially expressed genes, we constructed a PPI network, and we found the hub gene, PTGS2, which is an inducible isoenzyme (COX-2) of PTGS that causes colonic dysfunction after obstruction. Studies have confirmed that expression of COX-2 mRNA is significantly upregulated when the contractile ability of the intestinal smooth muscle expands proximal to the obstruction. Moreover, contractile ability is restored with a COX-2 selective inhibitor.24 Athanasios Tsigaridas et al performed proteomic analysis on serum samples of 30 irritable bowel syndrome patients and 10 healthy individuals, identifying eight differentially expressed proteins, including APOE, a polymorphic protein; in subgroup analysis, it was found that APOE was highly expressed in the diarrhea group.25 Ryotaro Ogawa et al found that CXCL8 correlates highly with angiogenesis and progression of colorectal cancer. High expression of CXCL8 in colorectal cancer leads to a very poor prognosis and promotes liver metastasis. Many proteins promote the proliferation of colorectal cancer cells through the CXCL1/8-CXCR2 axis.26
Whether STC and colorectal cancer are related is worthy of discussion. Most scholars believe that there is no obvious causal relationship and that STC is a functional disease not associated with malignant transformation and that colorectal cancer is related to gene mutation and lifestyle. Eun Mi Song et al confirmed that constipation does not significantly increase the incidence of colorectal cancer.27 Although constipation does not appear to increase the incidence of colorectal cancer, some research results confirm an increase in the incidence of polyps, which are precancerous lesions. One retrospective study showed that the incidence of colorectal cancer in elderly patients with constipation over 60 years old was higher than that in patients under 60 years old. The incidence of colorectal cancer in patients with constipation durations greater than 5 years is higher than that in patients with constipation durations less than 5 years. In the next in-depth analysis, we intersected the differential genes of STC patients with GSE113513 and GSE146587, and found that AQP8 and CFD may be the common risk factors of STC and CRC, and it is very likely to promote the progression of STC to CRC.
AQP8 is a member of the aquaporin family, the members of which act as water channels. Based on immunohistochemistry, AQP8 is mainly located in colonic absorptive epithelial cells; it plays an important role in the metabolism of colonic water.28 Expression of AQP8 mRNA in the ascending colon of patients with constipation is lower than that in the descending colon, whereas expression of AQP8 mRNA in the descending colon of patients with constipation is higher than that in controls. It has been suggested that a change in AQP8 expression is related to the occurrence of constipation. After AQP8 expression in the descending colon of patients with constipation increases, water absorption increases, resulting in dry stool. A study by Teng-Gen Hu et al on treatment of constipation with mulberry found that compared with the constipation group, expression of AQP8 in the mulberry treatment group was reduced but that the serum excitatory neurotransmitter level was increased. After mulberry treatment, the abundance of colon mucous cells increased, as did the number of lactobacilli and Bifidobacteria.
AQP8 is abnormally expressed in a variety of tumors. Wu et al found that AQP8 overexpression inhibits the tumorigenic phenotype by blocking PI3K/AKT signaling and inhibiting PCDH7 expression, thereby reducing the growth, invasiveness and colony formation of the colorectal cancer cell line HT-29.29 Using bioinformatics analysis, Xu et al reported that low expression of AQP8 indicates a poor prognosis in colorectal cancer.30 Zhang et al also confirmed that AQP8 is a direct target of miR-92a. miR-92a promotes the growth and invasion of CRC cells by negatively regulating expression of AQP8.31 In our analysis, AQP8 was found to have an effect on overall survival in patients with colorectal cancer. AQP8 plays an extremely important role in the occurrence and development of constipation and colorectal cancer. Long-term use of nonfibrous laxatives by patients with STC increases the incidence of colon polyps, which may be related to mechanical stimulation of the intestinal wall by dry stool. Colon polyps are a risk factor for cancer. Although we cannot directly prove a correlation between STC and colorectal cancer, our findings provide a direction for our future research.
Complement is a protein response system composed of proteins widely present on the surface of serum, tissue fluid and cell membranes with a precise regulatory mechanism. Complement is widely involved in the body’s microbial defense response and immune regulation, and it can also mediate the injurious effects of immune system pathology.32 The complement system involves nearly 40 components, most of which are glycoproteins. Complement C3 is the most abundant component in the complement system, and complement factor D has the smallest molecular weight. It has been reported that complement factor C3 mRNA is broadly present in intestinal mucosal cells in the lower part of the crypt.33 However, there are very few reports about the relationship between complement factor D and colorectal diseases. Our study also found no effect of CFD on overall survival in patients with colorectal cancer.
In a retrospective study, A Guérin found that patients with chronic constipation had significantly higher prevalence and incidence of colorectal cancer than matched patients with chronic non-constipation. These risks increase with the severity of chronic constipation.34 In a prospective cohort study of 75,214 participants in the United States, the risk of colorectal cancer was found to increase with the use of non-fibrous laxatives and decrease with the use of fibrous laxatives.14 The relationship between constipation and colorectal cancer has been debated, but the above research shows that there is indeed a link between the two, and constipation seems to increase the risk of colorectal cancer. This also attracted our attention even more.
Conclusion
We are the first to attempt to uncover the real cause of constipation at the genetic level by performing high-throughput sequencing of tissues from STC patients and normal individuals. At the same time, it was found that APQ8 may be a key gene marker that promotes the development of CRC in patients with constipation and affects the survival time. It has the potential to non-invasively screen high-risk groups of CRC and predict prognosis, and has great clinical significance. Of course, this study has certain limitations. First, few samples were used for RNA sequencing. Second, RCP verification was not performed on the hub genes. Finally, the pathological mechanism of STC should be confirmed by in vivo and in vitro experiments.
Data Sharing Statement
The datasets generated during and/or analysed during the current study are available in the SRA repository, [https://dataview.ncbi.nlm.nih.gov/object/PRJNA751889].
Ethics Approval and Informed Consent
The study was approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University. The procedures used in this study adhere to the tenets of the Declaration of Helsinki. For this study, we have obtained patient consent and signed an informed consent form.
Consent for Publication
Participants consent to this study published in a journal study.
Acknowledgments
Here, I would like to express my gratitude to all those who helped me during the writing process of my thesis. First of all, I would like to thank the experts for their guidance. Your professional and detailed opinions make our research more rigorous. Secondly, I would like to express my sincere thanks to my friends and classmates, who gave me help and time during the difficult process of the thesis, listened to my opinions, and helped me solve the problem.
Author Contributions
All authors made a significant contribution to the work reported, in the conception, study design, execution, acquisition of data, analysis and interpretation all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (No. 31011180065), the fund for scientific research from Pfizer, China (No. WI216584) and Natural Science Foundation of Heilongjiang Province of China (YQ2019H018).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Black CJ, Ford AC. Chronic idiopathic constipation in adults: epidemiology, pathophysiology, diagnosis and clinical management. Med J Aust. 2018;209(2):86–91. doi:10.5694/mja18.00241
2. Camilleri M, Ford AC, Mawe GM, et al. Chronic constipation. Nat Rev Dis Primers. 2017;3:17095. doi:10.1038/nrdp.2017.95
3. Drossman DA, Li Z, Andruzzi E, et al. U.S. householder survey of functional gastrointestinal disorders. Prevalence, sociodemography, and health impact. Dig Dis Sci. 1993;38(9):1569–1580. doi:10.1007/BF01303162
4. Walia R, Mahajan L, Steffen R. Recent advances in chronic constipation. Curr Opin Pediatr. 2009;21(5):661–666. doi:10.1097/MOP.0b013e32832ff241
5. Spiller R, Camilleri M, Longstreth GF. Do the symptom-based, Rome criteria of irritable bowel syndrome lead to better diagnosis and treatment outcomes? Clin Gastroenterol Hepatol. 2010;8(2):125–129;discussion 129–136. doi:10.1016/j.cgh.2009.12.018
6. Schiller LR. Chronic constipation: new insights, better outcomes? Lancet Gastroenterol Hepatol. 2019;4(11):873–882. doi:10.1016/S2468-1253(19)30199-2
7. Zhao S, Chen Q, Kang X, Kong B, Wang Z. Aberrantly expressed genes and miRNAs in slow transit constipation based on RNA-seq analysis. Biomed Res Int. 2018;2018:2617432. doi:10.1155/2018/2617432
8. Yan S, Yue YZ, Wang XP, et al. Aqueous extracts of herba cistanche promoted intestinal motility in loperamide-induced constipation rats by ameliorating the interstitial cells of cajal. Evid Based Complement Alternat Med. 2017;2017:6236904. doi:10.1155/2017/6236904
9. Israelyan N, Del Colle A, Li Z, et al. Effects of serotonin and slow-release 5-hydroxytryptophan on gastrointestinal motility in a mouse model of depression. Gastroenterology. 2019;157(2):507–521 e504. doi:10.1053/j.gastro.2019.04.022
10. Giancola F, Torresan F, Repossi R, et al. Downregulation of neuronal vasoactive intestinal polypeptide in Parkinson’s disease and chronic constipation. Neurogastroenterol Motil. 2017;29(5):e12995. doi:10.1111/nmo.12995
11. Tian Y, Wang L, Ye JW, et al. Defecation function and quality of life in patients with slow-transit constipation after colectomy. World J Clin Cases. 2020;8(10):1897–1907. doi:10.12998/wjcc.v8.i10.1897
12. Song EM, Lee HJ, Jung KW, et al. Long-term risks of parkinson’s disease, surgery, and colorectal cancer in patients with slow-transit constipation. Clin Gastroenterol Hepatol. 2021;19(12):2577–2586 e2576. doi:10.1016/j.cgh.2020.08.059
13. Simons CC, Schouten LJ, Weijenberg MP, Goldbohm RA, van den Brandt PA. Bowel movement and constipation frequencies and the risk of colorectal cancer among men in the Netherlands cohort study on diet and cancer. Am J Epidemiol. 2010;172(12):1404–1414. doi:10.1093/aje/kwq307
14. Citronberg J, Kantor ED, Potter JD, White E. A prospective study of the effect of bowel movement frequency, constipation, and laxative use on colorectal cancer risk. Am J Gastroenterol. 2014;109(10):1640–1649. doi:10.1038/ajg.2014.233
15. Rao SS, Rattanakovit K, Patcharatrakul T. Diagnosis and management of chronic constipation in adults. Nat Rev Gastroenterol Hepatol. 2016;13(5):295–305. doi:10.1038/nrgastro.2016.53
16. Xie XY, Sun KL, Chen WH, et al. Surgical outcomes of subtotal colectomy with antiperistaltic caecorectal anastomosis vs total colectomy with ileorectal anastomosis for intractable slow-transit constipation. Gastroenterol Rep. 2019;7(6):449–454. doi:10.1093/gastro/goz014
17. Guerin A, Carson RT, Lewis B, Yin D, Kaminsky M, Wu E. The economic burden of treatment failure amongst patients with irritable bowel syndrome with constipation or chronic constipation: a retrospective analysis of a Medicaid population. J Med Econ. 2014;17(8):577–586. doi:10.3111/13696998.2014.919926
18. Wolf PG, Parthasarathy G, Chen J, et al. Assessing the colonic microbiome, hydrogenogenic and hydrogenotrophic genes, transit and breath methane in constipation. Neurogastroenterol Motil. 2017;29(10):1–9. doi:10.1111/nmo.13056
19. Drossman DA. Functional gastrointestinal disorders: history, pathophysiology, clinical features and Rome IV. Gastroenterology. 2016;150(6):1262–1279.e2. doi:10.1053/j.gastro.2016.02.032
20. Mazzone A, Strege PR, Gibbons SJ, et al. microRNA overexpression in slow transit constipation leads to reduced NaV1.5 current and altered smooth muscle contractility. Gut. 2020;69(5):868–876. doi:10.1136/gutjnl-2019-318747
21. Wang Y, Wang Q, Kuerban K, et al. Colonic electrical stimulation promotes colonic motility through regeneration of myenteric plexus neurons in slow transit constipation beagles. Biosci Rep. 2019;39(5):BSR20182405.
22. Cheng L, Pricolo V, Biancani P, Behar J. Overexpression of progesterone receptor B increases sensitivity of human colon muscle cells to progesterone. Am J Physiol Gastrointest Liver Physiol. 2008;295(3):G493–502. doi:10.1152/ajpgi.90214.2008
23. Zhan Y, Tang X, Xu H, Tang S. Maren pills improve constipation via regulating AQP3 and NF-kappaB signaling pathway in slow transit constipation in vitro and in vivo. Evid Based Complement Alternat Med. 2020;2020:9837384. doi:10.1155/2020/9837384
24. Shi XZ, Lin YM, Powell DW, Sarna SK. Pathophysiology of motility dysfunction in bowel obstruction: role of stretch-induced COX-2. Am J Physiol Gastrointest Liver Physiol. 2011;300(1):G99–G108. doi:10.1152/ajpgi.00379.2010
25. Tsigaridas A, Anagnostopoulos AK, Papadopoulou A, et al. Identification of serum proteome signature of irritable bowel syndrome: potential utility of the tool for early diagnosis and patient’s stratification. J Proteomics. 2018;188:167–172. doi:10.1016/j.jprot.2017.07.019
26. Xiao YC, Yang ZB, Cheng XS, et al. CXCL8, overexpressed in colorectal cancer, enhances the resistance of colorectal cancer cells to anoikis. Cancer Lett. 2015;361(1):22–32. doi:10.1016/j.canlet.2015.02.021
27. Song EM, Lee HJ, Jung KW, et al. Long-term risks of parkinson’s disease, surgery, and colorectal cancer in patients with slow-transit constipation. Clin Gastroenterol Hepatol. 2020;19(12):2577–2586.e6.
28. Dotti I, Mora-Buch R, Ferrer-Picon E, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66(12):2069–2079. doi:10.1136/gutjnl-2016-312609
29. Wu Q, Yang ZF, Wang KJ, et al. AQP8 inhibits colorectal cancer growth and metastasis by down-regulating PI3K/AKT signaling and PCDH7 expression. Am J Cancer Res. 2018;8(2):266–279.
30. Xu H, Ma Y, Zhang J, et al. Identification and verification of core genes in colorectal cancer. Biomed Res Int. 2020;2020:8082697. doi:10.1155/2020/8082697
31. Zhang H, Du WB, Guo XM, Wang LK, Cheng JM, Wei LJ. Identification of the AQP8-miR-92a network associated with the aggressive traits of colorectal cancer. Biochem Biophys Res Commun. 2020;527(1):218–225. doi:10.1016/j.bbrc.2020.04.055
32. Beitia M, Romano P, Larrinaga G, et al. The activation of prothrombin seems to play an earlier role than the complement system in the progression of colorectal cancer: a mass spectrometry evaluation. Diagnostics. 2020;10(12):1077. doi:10.3390/diagnostics10121077
33. Liang JZ, Liang XL, Zhong LY, Wu CT, Zhang J, Wang Y. Comparative proteome identifies complement component 3-mediated immune response as key difference of colon adenocarcinoma and rectal adenocarcinoma. Front Oncol. 2020;10:617890. doi:10.3389/fonc.2020.617890
34. Guerin A, Mody R, Fok B, et al. Risk of developing colorectal cancer and benign colorectal neoplasm in patients with chronic constipation. Aliment Pharmacol Ther. 2014;40(1):83–92. doi:10.1111/apt.12789
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.