Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 13
Analysis of Comprehensive Pharmacogenomic Profiling of VIP Variants Among the Genetically Isolated Chechen Subpopulation from Jordan
Authors AL-Eitan LN , Rababa'h DM, Hakooz NM, Alghamdi MA , Dajani RB
Received 20 March 2020
Accepted for publication 29 June 2020
Published 14 July 2020 Volume 2020:13 Pages 199—215
DOI https://doi.org/10.2147/PGPM.S254677
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Laith N AL-Eitan,1,2 Doaa M Rababa’h,1 Nancy M Hakooz,3 Mansour A Alghamdi,4,5 Rana B Dajani6– 8
1Department of Applied Biological Sciences, Jordan University of Science and Technology, Irbid, Jordan; 2Department of Biotechnology and Genetic Engineering, Jordan University of Science and Technology, Irbid, Jordan; 3Department of Biopharmaceutics and Clinical Pharmacy, School of Pharmacy, University of Jordan, Amman, Jordan; 4Department of Anatomy, College of Medicine, King Khalid University, Abha, Saudi Arabia; 5Genomics and Personalized Medicine Unit, College of Medicine, King Khalid University, Abha, Saudi Arabia; 6Department of Biology and Biotechnology, Hashemite University, Zarqa, Jordan; 7Radcliffe Institute for Advanced Studies, Harvard University, Cambridge, MA, USA; 8Jepson School of Leadership, Richmond University, Richmond, VA, USA
Correspondence: Laith N AL-Eitan
Department of Applied Biological Sciences, Jordan University of Science and Technology, PO Box 3030, Irbid 22110, Jordan
Tel +962 -2 -7201000
Fax +962-2-7201071
Email [email protected]
Background: Profiling rare variants in isolated populations can significantly clarify and understand the development of a clinically relevant process. Therefore, leading to a better identifying novel targeted treatment.
Objective: This study aimed to determine the allele frequencies of 56 single nucleotide polymorphisms (SNPs) within several important pharmacogenes.
Methods: This study consisted of 166 unrelated subjects from a genetically isolated group (Chechen) who were living in Jordan. In this study, the distribution of the variants among Chechen was compared to other ethnic groups available at two databases (Genome 1000 and (ExAC)). The frequency of genotypes and alleles was calculated and tested using the chi-square test and the Hardy–Weinberg equilibrium equation (HWE).
Results: Our results revealed that the distribution of allele frequencies within different pharmacogenes among Chechen showed different similarities with other populations. The CEU and TSI showed the highest resemblance with the Chechen population (75% similarity), in contrast to LWK which had the lowest similarity (30%).
Conclusion: This study sheds light on clinically relevant SNPs to enhance medical research and apply pharmacogenomics in clinical settings.
Keywords: pharmacogenomics, VIP polymorphism, Phase I enzymes, Phase II enzymes, metabolizing
Introduction
Pharmacogenomics has become a promising future to enhance individuals’ response to treatment.1 Single nucleotide polymorphisms (SNPs) are one of the most common variants among populations that affect enzyme activity as well as drug metabolism.2 Consequently, clinical significant SNPs have been studied and screened extensively in different genes in different ethnic backgrounds and given the term very important pharmacogenomic (VIP) variants.3
These VIP variants help in drug dose optimization for patients relevant to their genetic makeup and lead to enhanced therapy outcomes and maintain the efficacy of drug treatment.4 Remarkably, the frequency distribution of these VIP variants varies among different ethnic groups.5 The importance of the VIP variants depends on their presence within critical genes including drug-metabolizing genes.6 However, the underlying mechanism of the correlation between the genetic variants and drug response is still beyond clinical practice.7 Rare variants that impact gene function may account for some uncovered differences in the pharmacological response and metabolism process.8 Genetic variants in drug target enzymes; metabolizing, transporters, receptors help in predicting toxicity and treatment failure among patients.9
Genetically isolated populations, such as Chechen, can help medical research to overcome the extensive allelic heterogeneity which is a major cause of complex diseases.10 Chechen, a Caucasian ethnic group of the Nakh peoples, lived in the highlands of the North Caucasus region of Eastern Europe.11 In 1860 they immigrated to Ottoman lands; however, today, tens of thousands of Chechens live in Turkey, Jordan, Syria, Iraq, Egypt, Saudi Arabia and other Persian Gulf countries.12 Not many genetic studies have been conducted on Chechens and Circassians living in Jordan. As a genetically isolated population, they can provide a baseline for future studies to discover novel markers associated with diseases.13–16 Moreover, there is an urgent need for pharmacogenetic research as not many studies conducted in Jordan.17,18 Therefore, this study aimed to determine the allele frequencies of 52 SNPs within several pharmacogenes in Chechen living in Jordan in comparison to other ethnic groups.
Materials and Methods
Study Subjects
This study was subjected to and in agreement with the Human ethics committee at National Center for Diabetes, Endocrinology and Genetics (NCDEG) and Jordan University of Science and Technology (JUST). Details about subjects and written informed consent were obtained from all volunteers in the study. Ten milliliters of venous blood was collected from unrelated healthy 166 Chechen subjects living in Jordan.
This study was conducted in agreement with the Human ethics committee at National Center for Diabetes, Endocrinology and Genetics (NCDEG) and Jordan University of Science and Technology (JUST), policy number (GM7601). Written informed consent was obtained from all volunteers in the study. This research was also conducted in accordance with the Declaration of Helsinki.
DNA Extraction and Genotyping
Blood samples were drawn from subject and then stored at 4°C for 24 hours before the purification of the genetic material. The extracted DNA was then stored at −20°C.
Genotyping was performed using Agena Bioscience MassARRAY® on a Compact Spectrometer iPLEX GOLD chemistry by Australian Genome Research Facility. Genomic DNA was extracted from each blood sample using the Wizard® Genomic DNA Purification Kit (Promega Corporation, USA) according to the manufacturer’s instructions. The quality and quantity of the purified DNA were ascertained via agarose gel electrophoresis and the Nano-Drop ND-1000 UV-Vis Spectrophotometer (BioDrop, UK), respectively.
Candidate genes polymorphisms were analyzed using Sequencing technique. Loci of candidate SNPs were amplified using Multiplex PCR followed by Mass EXTEND which is a primer extension process is resulting in allele-specific DNA products. The genotyping after the minisequencing reaction product analysis was reported by Mass spectrometry. After that, the extension PCR products were separated onto a 384 well spectroCHIP and placed into the MALDI-TOF (Matrix-Assisted Laser Desorption/Ionization Time-of-Flight) mass spectrometer. Lastly, a software system (SpectroTYPER-RT (RT for real-time)) was used to analyze the results.
Variants Selection
The studied VIP variants were selected from the public databases including the PharmGKB database (https://www.pharmgkb.org/), the SNP database of the National Centre for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/SNP/), and the International HapMap Project (http://www.hapmap.org/). SNPs selected within significant genes that play roles in drug metabolism which make their variants clinically important variants. Finally, the primers were used to genotype these VIP variants were shown in the Supplementary Material.
Population’s Variation Data
The allele counts variation data for other populations were obtained from HapMap Project website: CHE: Chechens from Jordan, ASW: African ancestry in Southwest USA, CEU: Utah, USA residents with Northern and Western European ancestry from the CEPH collection, CHB: Han Chinese in Beijing, China, CDX: Chinese Dai in Xishuangbanna, China, GIH: Gujarati Indians in Houston, Texas, USA, GBR: British in England and Scotland, JPT: Japanese in Tokyo, Japan, LWK: Luhya in Webuye, Kenya, MXL: Mexican ancestry in Los Angeles, California, USA, TSI: Toscani in Italy, YRI: Yoruba in Ibadan, Nigeria, CAR: Circassian from Jordan (unpublished data), ACB; African Caribbeans in Barbados. Also, the allele count data of populations around the world were obtained from the Exome Aggregation Consortium (ExAC): African, East Asian, Latino, European (non-Finnish), South Asian and European (Finnish).
Statistical Analysis
Statistical Package for Social Sciences SPSS (version 19) was used to statistically analyze the data. The genotypes and alleles frequency was calculated and tested using the chi-square test and the Hardy–Weinberg equilibrium equation (HWE) and all p-values accepted as p-value <0.05. Fisher exact test was used for the correction of small sizes.
Results
Table 1 shows basic information about the selected variants within 28 studied genes, in addition to describe the quality control of samples’ genotyping. Genotype call rates ranged from 96.5% to 100%. Allele and genotype frequencies of the selected variants among 166 Chechen subjects are listed in Table 2. SNPs were tested for Hardy–Weinberg Equilibrium (HWE), all the studied polymorphisms met the HWE standards (P-value > 0.05) except for rs1801131 within MTHFR (P-value = 0.027) did not fulfil the HWE equation (Figure 1).
 |
Table 1 List of Genes, Their VIP Variants, Positions, and Genotyping Data Based on the Whole Cohort (N= 166) |
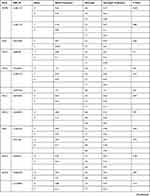 |  |  |  |
Table 2 The Minor Allele Frequencies and HWE p Values for the VIP Variants in Chechen (N= 166) |
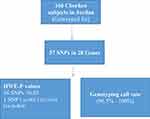 |
Figure 1 The number of single nucleotide polymorphisms (SNPs) in this study and their genotyping accuracy. |
Table 3 compares minor allele frequencies (MAF) of the significant polymorphisms among Chechens and other populations. For example, the MAF of rs776746 (T) of the CYP3A5 gene within the Chechen group was similar to both CEU and GBR but different from ASW, CHB, CDX, GIH, and others. Also, Table 3 compares the allelic distribution of selected variants among Chechens and Circassians and other populations by estimating the P-value. We found that each studied variant distribution within Chechens in Jordan is significantly different from various populations. For example, Rs1229984 and rs28371725 had a highly different distribution with around 90% of the compared populations.
 |  |  |  |
Table 3 VIP Variants Within the Pharmacogenes in Chechen Compared to HapMap Populations |
It is also clear from Table 3 that the similarity of the studied polymorphisms within the Chechens population with other compared population varied from one to another. The CEU Utah (USA residents with Northern and Western European ancestry from the CEPH collection) and TSI (Toscani in Italy) had the highest similarity among the studied population with around 25% differences; ie, around 75% of the studied variants had the same frequency and distribution in Chechens and other compared populations. This similarity in the distribution of the compared SNPs decreased to reach around 30% when compared to LWK (Luhya in Webeye).
Furthermore, the significant variants within the selected genes within Chechen were compared to the available variants within six populations listed in the Exome Aggregation Consortium (ExAC) database as shown in Table 4.
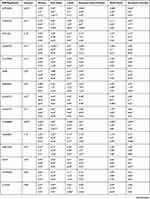 | 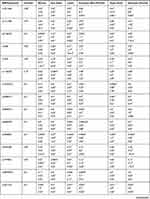 |  |
Table 4 VIP Variants Within the Pharmacogenes in Chechen Compared to Six ExAC Populations Worldwide |
Discussion
This study was conducted on Chechens group living in Jordan. This subpopulation has a distinct genetic makeup as a genetically isolated population.19 Our findings demonstrate that the Chechen living in Jordan may have widely varying genetic allele distribution for clinically relevant SNPs compared to other populations. Assessment of the VIP genetic polymorphism frequencies within such a minority group can attribute to studies of the theoretical basis of drug toxicity and efficacy.
Several single nucleotide variants within the CYP450 genes have been reported as clinically significant. For example; within the CYP2J2 gene, the variant rs890293 genotypes AA + AC have been shown to be correlated to an increased risk of nausea and vomiting when treated with tacrolimus in individuals with Kidney Transplantation as compared to genotype CC.20
rs4244285 (GG) genotype of CYP2C19 is associated with increased risk of toxicity when treated with cyclophosphamide in women with Lupus Erythematosus, as compared to genotypes AA + AG.21 Besides, patients with the genotype GG of rs4986893 and treated with phenytoin may have a decreased likelihood of drug reaction with eosinophilia and systemic symptoms (DRESS) as compared to patients with the AG and AA genotypes.22 It also was found that allele A is associated with an increased risk of cardiovascular events when treated with clopidogrel in patients with coronary disease as compared to genotype GG.23 Moreover, allele A of rs4986893 is associated with decreased clopidogrel inhibition of ADP-induced platelet aggregation when treated with clopidogrel in healthy individuals as compared to genotype GG.24,25
Furthermore, our findings reveal that the alternative allele (T) frequency of rs1229984 in ADH1B gene among Chechen was much less than in Latino (94%), African (99%), European non-Finnish (95%), South Asian (95%) and European Finnish (99%) according to ExAC database. rs1229984 was reported as a clinically significant variant, whereas TT genotype was associated with increased Vmax of ethanol in healthy individuals as compared to genotypes CC and CT among mixed population.26
Furthermore, within ADRB2 gene, the frequency of the minor allele (G) of rs1042714 among Chechen was significantly lower than it among others Latino (83%), African 82%, European non-Finnish (58%), South Asian 79 and European Finnish (63%) and East Asian (91%). It was suggested that allele G of rs1042714 was associated with an increased reduction in resting blood pressure when treated with carvedilol in healthy individuals as compared to allele G in a study conducted in a mixed population.27 While Genotype CC was reported to be associated with decreased improvement in heart failure patients when treated with carvedilol compared to genotypes GG and CG.28
In this study, we found that rs5219 of KCNJ11 gene among Chechen was distributed differently from other populations. It was proposed that the Genotype (TT) is associated with an increased likelihood of Diabetes II Mellitus.29
One significant limitation for this study is the sample size, large study cohort is recommended to compare the variants frequencies with other population. In addition, better designed statistical analyses should be provided for accurate results. In conclusion, rare variants detected in isolated populations can significantly guide to understanding a potential biological process and identifying genetic loci involved in the development of clinically relevant treatment for different diseases.
Ethics Committee Approval and Patient Consent
This study was conducted in agreement with the Human ethics committee at National Center for Diabetes, Endocrinology and Genetics (NCDEG) and Jordan University of Science and Technology (JUST), policy number (GM7601). Written informed consent was obtained from all volunteers in the study.
Disclosure
The authors declare that they have no competing interests.
References
1. Burroughs VJ, Maxey RW, Levy RA. Racial and ethnic differences in response to medicines: towards individualized pharmaceutical treatment. J Natl Med Assoc. 2002;94:1–26.
2. Brumfield RT, Beerli P, Nickerson DA, et al. The utility of single nucleotide polymorphisms in inferences of population history. Trends Ecol Evol. 2003;18:249–256.
3. He Y, Yang H, Geng T, et al. Genetic polymorphisms of pharmacogenomics VIP variants in the Ihoba population of southwest China. Int J Clin Exp Pathol. 2015;8:13293–13303.
4. Nebert DW, Menon AG. Pharmacogenomics, ethnicity, and susceptibility genes. Pharmacogenomics J. 2001;1:19–22.
5. Pennisi E. Breakthrough of the year. Human genetic variation. Science. 2007;318:1842–1843. doi:10.1126/science.318.5858.1842
6. Suh Y, Cantor C. Single nucleotide polymorphisms (SNPs): detection, interpretation, and application. Mutat Res. 2005;573:1–2. doi:10.1016/j.mrfmmm.2005.01.003
7. Altshuler DM, Gibbs RA, et al.; International HapMap Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58.
8. Bush WS, Crosslin DR, Owusu-Obeng A, et al. Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin Pharmacol Ther. 2016;100(2):160–169. doi:10.1002/cpt.350
9. Engen RM, Marsh S, Van Booven DJ, McLeod HL. Ethnic differences in pharmacogenetically relevant genes. Curr Drug Targets. 2006;7:1641–1648. doi:10.2174/138945006779025446
10. Kristiansson K, Naukkarinen J, Peltonen L. Isolated populations and complex disease gene identification. Genome Biol. 2008;9:109–117.
11. Anchabadze G. The Vainakhs (The Chechen and Ingush).
12. Jaimoukha A. The Chechens: A Hand Book.
13. Elbein SC. Genetics factors contributing to type 2 diabetes across ethnicities. J Diabetes Sci Technol. 2009;3(4):685–689. doi:10.1177/193229680900300412
14. Dajani R, Fatahallah R, Dajani A, Al-Shboul M, Khader Y. Prevalence of coagulation factor II G20210A and factor V G1691A Leiden polymorphisms in Chechans, a genetically isolated population in Jordan. Mol Biol Rep. 2012;39(9):9133–9138. doi:10.1007/s11033-012-1785-7
15. AL-Eitan L, Nassar A, Dajani R, Almomani B, Saadeh N. Diabetes mellitus in two genetically distinct populations in Jordan. A Comparison between Arabs and Circassians/Chechens Living with Diabetes. Saudi Med J. 2017;38(2):163–169.
16. AL-Eitan L, Mohammad N, Al-Maqableh H, Hakooz N, Dajani R. Genetic Polymorphisms of Pharmacogenomic VIP Variants in the Circassian Subpopulation from Jordan. Curr Drug Metab. 2019;20(8):674–681. doi:10.2174/1389200220666190729124000
17. AL-Eitan L, Tarkhan AH. Practical challenges and translational issues in pharmacogenomics and personalized medicine from 2010 onwards. Curr Pharmacogenomics Person Med. 2016;14:7–17.
18. AL-Eitan L, Haddad YA. Emergence of pharmacogenomics in academic medicine and public health in jordan: history, present state and prospects. Curr Pharmacogenomics Person Med. 2014;12:167–175.
19. Hakooz N, Alzubiedi S, Yousef AM, et al. UDP-glucuronosyltransferase 1A4 (UGT1A4) polymorphisms in a Jordanian population. Mol Biol Rep. 2012;39(7):7763–7768. doi:10.1007/s11033-012-1615-y
20. Genvigir FD, Nishikawa AM, Felipe CR, et al. Influence of ABCC2, CYP2C8, and CYP2J2 polymorphisms on tacrolimus and mycophenolate sodium-based treatment in brazilian kidney transplant recipients. Pharmacotherapy. 2017;37:535–545.
21. Ngamjanyaporn P, Thakkinstian A, Verasertniyom O, et al. Pharmacogenetics of cyclophosphamide and CYP2C19 polymorphism in Thai systemic lupus erythematosus. Rheumatol Int. 2011;31:1215–1218. doi:10.1007/s00296-010-1420-7
22. Yampayon K, Sukasem C, Limwongse C, et al. Influence of genetic and non-genetic factors on phenytoin-induced severe cutaneous adverse drug reactions. Eur J Clin Pharmacol. 2017;73:855–865. doi:10.1007/s00228-017-2250-2
23. Yamamoto K, Hokimoto S, Chitose T, et al. Impact of CYP2C19 polymorphism on residual platelet reactivity in patients with coronary heart disease during antiplatelet therapy. J Cardiol. 2011;57:194–201. doi:10.1016/j.jjcc.2010.10.007
24. Chen BL, Zhang W, Li Q, et al. Inhibition of ADP-induced platelet aggregation by clopidogrel is related to CYP2C19 genetic polymorphisms. Clin Exp Pharmacol Physiol. 2008;35:904–908.
25. Schroth W, Antoniadou L, Fritz P, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–5193. doi:10.1200/JCO.2007.12.2705
26. Seng KY, Limenta LM, Heng D, Lee EJ. Population pharmacokinetics and pharmacogenetics of alcohol in Chinese and Indians in Singapore. J Clin Pharm Ther. 2013;38:141–149. doi:10.1111/jcpt.12003
27. Sehrt D, Meineke I, Tzvetkov M, Gültepe S, Brockmöller J. Carvedilol pharmacokinetics and pharmacodynamics in relation to CYP2D6 and ADRB2 pharmacogenetics. Pharmacogenomics. 2011;12(6):783–795. doi:10.2217/pgs.11.20
28. Kaye DM, Smirk B, Williams C, Jennings G, Esler M, Holst D. Beta-adrenoceptor genotype influences the response to carvedilol in patients with congestive heart failure. Pharmacogenetics. 2013;13(7):379–382.
29. Gloyn AL, Hashim Y, Ashcroft SJ, Ashfield R, Wiltshire S, Turner RC. UK Prospective Diabetes Study (UKPDS 53). Association studies of variants in promoter and coding regions of beta-cell ATP-sensitive K-channel genes SUR1 and Kir6.2 with Type 2 diabetes mellitus (UKPDS 53). Diabet Med. 2001;18(3):206–212. doi:10.1046/j.1464-5491.2001.00449.x
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
