Back to Journals » Journal of Pain Research » Volume 11
Analgesic efficacy, safety, and tolerability of a long-acting abuse-deterrent formulation of oxycodone for moderate-to-severe chronic low back pain in subjects successfully switched from immediate-release oxycodone
Authors Markman J, Meske DS , Kopecky EA, Vaughn B, O'Connor ML, Passik SD
Received 21 March 2018
Accepted for publication 22 June 2018
Published 26 September 2018 Volume 2018:11 Pages 2051—2059
DOI https://doi.org/10.2147/JPR.S168836
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Michael Schatman
John Markman,1 Diana S Meske,2 Ernest A Kopecky,2 Ben Vaughn,3 Melinda L O’Connor,2 Steven D Passik2
1Department of Neurosurgery, Translational Pain Research Program, University of Rochester, Rochester, NY, USA; 2Collegium Pharmaceutical Inc., Canton, MA, USA; 3Rho Inc., Chapel Hill, NC, USA
Objectives: This post hoc analysis of data from a randomized, double-blind, placebo-controlled, enriched-enrollment randomized-withdrawal Phase III study evaluated the safety, tolerability, and analgesic efficacy of Oxycodone DETERx extended-release (ER), abuse-deterrent capsules (Xtampza® ER) in subjects with chronic low back pain who were successfully transitioned from immediate-release (IR) oxycodone.
Methods: Continuous outcomes were analyzed using a mixed-model repeated-measures approach; binomial outcomes were analyzed using chi-squared; and time-to-event outcomes using Kaplan–Meier analyses.
Results: A total of 110 subjects previously prescribed IR oxycodone entered the Open-label Titration Phase. Forty-four subjects were randomized to Oxycodone DETERx (n=22) or placebo (n=22) in the 12-week Double-blind Maintenance Phase. Efficacy results in this subgroup showed a statistically significant difference between Oxycodone DETERx and placebo in average pain intensity scores from Randomization Baseline to Week 12 (least squares mean [± standard error], –1.88 [0.70]; P=0.0078). Additional efficacy results indicated that Oxycodone DETERx vs placebo was associated with a statistically significant benefit in durability of effect from Week 2 through Week 12 (P<0.01), numbers of subjects with a ≥30% (n [%] 10 [45.5%] vs 0 [0%]; P=0.0004) and ≥50% (10 [45.5%] vs 0 [0%]; P=0.0004) improvement in pain intensity, longer time-to-exit (P=0.0014), a greater number of subjects who completed the study (14 [63.6%] vs 4 [18.2%]), and less rescue medication use (acetaminophen; mean [SD], 163.5 [337.8] mg) vs 216.2 [377.3] mg). Adverse event profiles were consistent with opioid class effects and results from the original study; Oxycodone DETERx was well tolerated in subjects previously treated with short-acting oxycodone.
Conclusions: Oxycodone DETERx resulted in clinically meaningful and statistically significant efficacy in subjects with chronic low back pain who were previously prescribed IR oxycodone and were successfully switched to ER Oxycodone DETERx.
Keywords: oxycodone, DETERx, Xtampza ER, chronic low back pain, extended-release, immediate-release, opioid
Background
The rapid increase in infections caused by antibiotic-resistant Gram-negative bacteria, such as carbapenem-resistant Enterobacteriaceae, has raised great concern worldwide. Colistin was used as an alternative therapy to treat infections caused by multidrug-resistant (MDR) Gram-negative bacteria.1 Colistin was one of the few options for such type of bacterial infections. As colistin is currently used for treating human or animal infections, an increasing number of colistin and drug resistance mechanisms are reported.2,3 Nevertheless, the prevalence of the colistin-resistant issue has gained worldwide attention. Mcr-1 genes were first reported as mediated by plasmid and can be spread between different bacteria through plasmids. Previously, the primary mechanism underlying colistin resistance stated that the bacterial chromosome coding of the binary regulatory system pmr AB and pho PQ and the associated regulatory gene mgr B mutations led to altered lipid A modification, thereby reducing the affinity of bacteria for polymyxin.4 In 2015, some investigators identified a novel mechanism mediating the low level of resistance to colistin.5 These resistant strains carry a novel gene mcr-1 encoding phosphoethanolamine transferase that reduces the affinity of colistin to lipopolysaccharide, which was identified on an IncI2 plasmid and pHNSHP45, isolated from an Escherichia coli isolate from a pig in China; thus, these bacteria are not deemed sensitive to colistin.6,7 Currently, the mcr-1 gene has been detected in approximately 40 countries. The distribution of these plasmids among carbapenem-resistant organisms might produce “super bacteria.”
Herein, we aimed to determine the prevalence of mcr-1 gene in a Chinese-teaching hospital and understand its molecular characteristics. Furthermore, we assessed the prognostic impact of this gene on clinical patients and the drug resistance of patients infected with mcr-1-positive strains. Hence, monitoring the resistance of colistin, delaying the spread of bacterial resistance, and providing effective advice for clinical treatment are essential.
Methods
Clinical bacterial isolates
The clinical isolates were collected from the Microbiology Laboratory of Anhui Medical University Union Hospital (Heifei, Anhui, China) from January 2015 to 2016, which is the largest tertiary hospital in Anhui Province, Central China. The inclusion criteria were distinct for the diagnosis of bacterial infections. All the isolates were identified using the Gram-negative bacteria identification card of the Vitek system (BioMèrieux, Missouri, France).
Extract template DNA
Several monoclonal bacterial colonies were suspended in 100 µL of sterile distilled water and heated at 100°C for 10 minutes and centrifuged at 10,000× g for 10 minutes. The resulting supernatant contained the bacterial genomic DNA.
mcr-1 screening
The primers for the PCR amplification of mcr-1 gene were as follows: mcr-1-forward (5′-GCTCGGTCAGTCCGTTTG-3′) and mcr-1-reverse (5′-GAATGCGGTGCGGTCTTT-3′). A total of 35 cycles were conducted as follows: 94°C predenaturation for 5 minutes, 94°C denaturation for 1 minute, 55°C annealed 30 seconds, and 72°C extension for 1 minute. The PCR products were analyzed by agarose gel electrophoresis (Figure 1), and the positive products were sequenced.
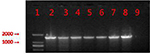  | Figure 1 PCR products from six mcr-1-positive isolates. Notes: 1: marker, DL2000; 2: positive control; 3: E4857; 4: E6512; 5: E0964; 6: E2069; 7: E9497; 8: E1825; and 9: negative control. |
Molecular typing
Pulsed-field gel electrophoresis (PFGE) was used for the molecular typing of six mcr-1-positive strains. The bacteria were embedded in SeaKem Gold Agarose (Lonza, Rockland, MD, USA), and the genome was subjected to restriction cleavage using XbaI. Subsequently, the DNA fragments were electrophoresed by the CHEF-mapper XA PFGE system (Bio-Rad Laboratories Inc., Hercules, CA, USA) for 22 hours at 14°C, 6 V/cm, and from 5 to 35 seconds pulses. The PFGE patterns were compared using BioNumerics Version 7.6. Furthermore, whole genome sequencing was used for determining the molecular typing and all the resistance genes.
mcr-1-resistant gene detection and analysis
The mcr-1 DNA was digested using the S1 nuclease (Takara, Otsu, Japan), and then, the genomic DNA resolved by PFGE system for 18 hours at 14°C with 2.16–63.8 seconds pulses. The DNA fragments were transferred to nylon membrane (EMD Millipore, Billerica, MA, USA) that was hybridized with digoxigenin-labeled mcr-1-specific probes in the hybridization oven at 40°C overnight. The NBT/BCIP color detection kit (Hoffman-La Roche Ltd., Basel, Switzerland) was used for staining the nylon membrane after hybridization.
We used filter mating to investigate whether the mcr-1-resistant gene can be transferred. E. coli EC600 with rifampicin resistance was employed as the recipient strain; all the mcr-1-positive isolates were tested, ratio of the donor and recipient strains was 1:1, and the agar with colistin (4 mg/L) and rifampicin (700 mg/L) was utilized for selection. The transconjugants were demonstrated as mcr-1 positive by PCR and further validated by PFGE. Then, the plasmids from mcr-1-positive transconjugants were extracted. Next, a strain of Klebsiella pneumoniae was preserved at the Department of Microbiology of our hospital. The growth did not produce mucus on the plate and resistance to imipenem; K. pneumoniae served as the recipient strain. The recipient bacteria were mixed with the extracted plasmid in a ratio of 10:1, and the electrotransformation mixture was set at a voltage of 2.1 kV. The electrotransformed mixture was incubated in the broth for 3 hours and allowed to grow overnight on plates containing polymyxin (2 mg/L).
Antimicrobial susceptibility testing (AST)
All mcr-1-positive specimens were tested for AST. The antimicrobial agents tested included meropenem, imipenem, tigecycline, colistin, aztreonam, amikacin, levofloxacin, cefoperazone-sulbactam, cefotaxime, cefepime, and trimethoprim/sulfamethoxazole. The Clinical and Laboratory Standards Institute (CLSI) was used to explain the results of AST.8 E-test was used for supplementing the susceptibility testing, and the results were interpreted based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints.9 E. coli 25922 was used as a standard quality control strain.
Gene sequence analysis
Positive strains were analyzed by whole-genome sequencing.
Clinical data collection
The clinical data including specimen type, age, sex, infection type, and clinical outcomes were collected from patients. These patients exhibited a clear infection during hospitalization. Also, they were treated with antibiotics during the hospital stay. The cases of these patients were analyzed retrospectively.
Results
In this study, we collected 1,112 nonduplicate clinical infection isolates from patients. Of these isolates, six (0.6%) strains carried the mcr-1 gene, which was similar to that reported previously.10–13 These mcr-1-positive isolates were collected from five different departments. The age of the patients ranged from 30 to 70 years, except for one of the patients, who was only 3 months old. The baseline data of the patients with mcr-1-positive E. coli infection are shown in Table 1. In addition, AST was estimated in all positive isolates and the MIC of colistin ranged from 4 to 16 mg/L, indicating a low level of resistance to polymyxin. Of the six mcr-1-positive isolates, a majority were susceptible to tigecycline, amikacin, cefoperazone sulbactam, meropenem, and imipenem, while only one was susceptible to levofloxacin, aztreonam, and trimethoprim/sulfamethoxazole, and all isolates were resistant to polymyxin, cefotaxime, and cefepime. The characteristics of mcr-1-positive isolates are shown in Tables 1–3.
  | Table 1 Clinical information of six patients with mcr-1-positive specimens |
Mcr-1 was usually found to be present in the bacterial genome with other resistance genes, such as ESBL.23 Nevertheless, all the six positive isolates were susceptible to carbapenems and meropenem. Moreover, the whole genome sequencing did not show any coexistence of carbapenem-resistant genes with mcr-1. We also found that the ESBL genes, including blaTEM-1B, blaCTX-M14, blaCTX-M132, blaCTX-M55, and blaCTX-M123 were contained in the genomes of these six strains that can lead to resistance to antibiotics, such as cephalosporins and penicillins. Notably, these strains contain other resistance genes: fosA3, fosA, aph(4)-Ia, sul2, oqxA, oqxB, and aadA2. A large number of studies have found that the transfer of the mcr-1 gene was usually associated with the movable element ISApl1-mcr-1. Our sequencing analysis did not reveal significant differences around the mcr-1 gene in the six mcr-1-positive strains, and two genetic environments were detected (Figure 2). We did not find the IsApl1 insert upstream to the mcr-1 gene, rather the pap-2 gene was found downstream to the mcr-1 gene. Thus, we speculated that the ISApl1 gene may be recombined, resulting in only partial sequences remaining upstream of mcr-1.
  | Figure 2 Two representativemcr-1 gene environments. |
XbaI-PFGE (Figure 3) showed that these mcr-1-positive strains were divided into six genotypes that indicated nonclonal transmission. The six mcr-1-positive strains were grouped into six distinct sequence types (STs) that were highly heterogeneous and a variety of plasmid type bacteria that carry mcr-1.
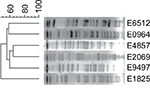  | Figure 3 Cluster analysis of six strains of bacteria by XbaI digestion. |
In the current filter mating study, two strains were successfully transferred to EC600 simultaneously, while the remaining four strains were tested two times, suggesting that all the mcr-1 genes were harbored in the plasmids. After 1 week of continuous subculture, the transconjugants can still detect the mcr-1 gene after PCR verification, which proves that the plasmid carrying mcr-1 can be stable in the transconjugants. In the plasmid conjugation test, the transconjugants did not grow on the plates, which might be attributed to the low efficiency of electrotransformation. This phenomenon might be related to the bacterial characteristics of K. pneumoniae.
The results of the S1 nuclease digestion (Figure 4) and Southern blot analysis (Figure 5) showed that the mcr-1 genes were localized on the following three different types of plasmids: approximately 33, 61, and >92 kb.
  | Figure 5 Southern blot hybridization of E. coli isolates with mcr-1. Notes: 1: E1825; 2: E9497; 3: E2069; 4: E0964; 5: E6512; and 6: E485. Abbreviation: E. coli, Escherichia coli. |
Discussion
The current study showed that the mcr-1 gene could be detected in various Enterobacteriaceae bacteria, including E. coli, K. pneumoniae, Salmonella, and Enterobacter cloacae, and reported in several countries.14–18 Some studies demonstrated that the mcr-1 gene could be transmitted through the food chain model, leading to its widespread occurrence in poultry farming, and sources of water contaminated with mcr-1 can also be considered as another route of transmission beyond the chain.5,12,19–21 The following two cases of mcr-1-positive E. coli were identified in the population: infected bacteria and fixed-value bacteria; the use of antibiotics and the patient’s immunosuppression are the putative causes of the epidemic.24 Currently, the detection of human Enterobacter mcr-1 has been reported in various countries at a rate of 1–2%, which is significantly lower than that of the animal sources.5,12 Usually, the mcr-1 gene coexists with other drug-resistant genes, which is in agreement with the current study. Other drug-resistant genes copromote mcr-1 that might indicate a selective role for mcr-1 and promote its epidemic spread; however, the underlying mechanism is not yet clarified. Based on the relevant research results, the lack of appropriate pressure selection did not prompt the occurrence of mcr-1 in the human body for a prolonged period.18
More than 10 plasmids have been found to carry the mcr-1 gene, including IncX4, Incl2, IncP, IncX3-IncX4, IncFII, Incl2-IncFIB, IncFI, IncX1-IncX2, IncHI1, and IncHI2. The most prevalent plasmids are IncHI2, Inc2, and Inc2, followed by IncX4 and other plasmids. The genome of these plasmids carries the mcr-1 gene, as well as, other drug-resistant genes, leading to the cotransmission of multiple drug-resistant genes. The insertion sequence ISApl1 is often detected upstream or downstream of the mcr-1 gene, and this mobile element causes the spread of mcr-1 gene. Some studies have shown that the mcr-1 gene can be inserted into other plasmids by its upstream and downstream looping in ISApl1.22
Nevertheless, the current study showed that the prevalence of colistin resistance was extremely low in E. coli based on the clinical infections in the hospital. This result was similar to that of the previous study.5,10–13 However, we need to further expand the number of strains and the sample collection areas. The diversity of PFGE types and STs showed that all of these mcr-1-positive isolates were from sporadic cases, and this low prevalence might be the background of the presence of the mcr-1 gene in microorganisms; however, additional data are essential for the substantiation of these findings. Hitherto, according to the results of Chinese studies, the mcr-1 gene has a high prevalence rate in animals and a low prevalence rate in hospital patients.10,25,26 Furthermore, in China, due to the large quantities of polymyxin used in agriculture and animal husbandry, the spread of mcr-1 gene between bacteria cannot be ignored. Although no mcr-1-positive K. pneumoniae was detected in the collected isolates in this study, a low level of mcr-1 prevalence was detected in K. pneumoniae.5,12,14–18 Considering that less clinical data are available for mcr-1, we aimed to establish an animal infection model to understand the correlation between the new resistance genes and other resistance genes and the threat to human infection.
mcr-1 can be transmitted between the natural environment, human body, and different strains through different transfer elements. Therefore, we can control the spread of mcr-1 by several effective measures. mcr-1 spreads in the hospital environment in the absence of colistin. Thus, patients with mcr-1 infection in the hospital, ward environment, medical equipment, as well as, doctors should be monitored promptly to administer antibiotics effectively in order to prevent the prevalence of mcr-1 in the hospital.
The major concern for the clinicians is the mcr-1 transfer into carbapenemase producers in a hospital setting, which could lead to the production of pandrug-resistant (PDR) strain.27,28 Thus, effective measures are imperative to monitor the prevalence of multislime resistance, thereby preventing the further spread of bacterial resistance.
Conclusion
The current study showed that the mcr-1 gene was responsible for the majority of colistin resistance in clinical isolates of E. coli. The gene can be transferred into plasmids containing other drug resistance genes by plasmid-DNA conjugation, which might cause severe consequences in drug-resistant strains. Thus, the widespread popularity of mcr-1 gene should be prevented.
Ethical approval
All patients provided written informed consent, and the Ethics approval was obtained from the medical ethics committee of the First Affiliated Hospital of Anhui Medical University with the following reference number: Quick-PJ 2018-05-14.
Acknowledgment
We thank Prof Yunsong Yu at the Affiliated Hospital of Zhejiang University, Zhejiang, for technical assistance during the experiment.
Author contributions
Prof Jilu Shen is responsible for the experimental design and is the corresponding author. Other authors are responsible for the experimental procedure and clinical data collection. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
None of the authors have any personal or financial involvement with the organizations that have financial interest in its content. The authors report no conflicts of interest in this work.
References
Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. | ||
Arduino SM, Quiroga MP, Ramírez MS, et al. Transposons and integrons in colistin-resistant clones of Klebsiella pneumoniae and Acinetobacter baumannii with epidemic or sporadic behaviour. J Med Microbiol. 2012;61(Pt 10):1417–1420. | ||
Gunn JS. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 2008;16(6):284–290. | ||
Falagas ME, Rafailidis PI, Matthaiou DK. Resistance to polymyxins: Mechanisms, frequency and treatment options. Drug Resist Updat. 2010;13(4-5):132–138. | ||
Lx Y, Liu YY, Rj W, et al. Plasmid - mediated colicin - resistant genes mcr-1 Research progress. Hereditas. 2017;39(2):110–126. | ||
Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295. | ||
Fernández L, Jenssen H, Bains M. The two-component system Cpr RS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of Par RS. Antimicrob Agents Chemother. 2012;56(12). | ||
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing[S]; 2016:M100–S26. | ||
EUCAST. Breakpoints Tables for Interpretation of MICs and Zone Diameters, Version 6.0; 2016. | ||
Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. | ||
Quan J, Li X, Chen Y, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–410. | ||
Hong XU, Xiao Ting LEI, Hua ZL, et al. Research progress on polymyxin resistance gene mcr-1. Chin J Nosocomiol. 2017;27(24):5741–5744. | ||
Zhong LL, Phan HTT, Shen C, et al. High rates of human fecal carriage of mcr-1-positive multi-drug resistant Enterobacteriaceae isolates emerge in China in association with succesful plasmid families. Clin Infect Dis. 2018;66(5):676–685. | ||
Webb HE, Granier SA, Marault M, et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(2):144–145. | ||
Tse H, Yuen K-Y. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(2):145–146. | ||
Hu Y, Liu F, Lin IYC, Gao GF, Zhu B. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(2):146–147. | ||
Olaitan AO, Chabou S, Okdah L, Morand S, Rolain JM. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(2):147. | ||
Arcilla MS, van Hattem JM, Matamoros S, et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(2):147–149. | ||
Liakopoulos A, Mevius DJ, Olsen B, Bonnedahl J. The colistin resistance mcr-1 gene is going wild. J Antimicrob Chemother. 2016;71(8):2335–2336. | ||
Ruzauskas M, Vaskeviciute L. Detection of the mcr-1 gene in Escherichia coli prevalent in the migratory bird species Larus argentatus. J Antimicrob Chemother. 2016;71(8):2333–2334. | ||
Mohsin M, Raza S, Roschanski N, Schaufler K, Guenther S. First description of plasmid-mediated colistin-resistant extended-spectrum β-lactamase-producing Escherichia coli in a wild migratory bird from Asia. Int J Antimicrob Agents. 2016;48(4):463–464. | ||
Snesrud E, He S, Chandler M, et al. A Model for Transposition of the Colistin Resistance Gene mcr-1 by ISApl1. Antimicrob Agents Chemother. 2016;60(11):6973–6976. | ||
Schwarz S, Johnson AP. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother. 2016;71(8):2066–2070. | ||
Wang Y, Tian GB, Zhang R, et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. 2017;17(4):390–399. | ||
Haenni M, Poirel L, Kieffer N, et al. Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis. 2016;16(3):281–282. | ||
Falgenhauer L, Waezsada SE, et alRESET consortium, et al. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis. 2016;16(3):282–283. | ||
Mulvey MR, Mataseje LF, Robertson J, et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(3):289–290. | ||
Guan X, He L, Chinese XDR Consensus Working Group. Laboratory diagnosis, clinical management and infection control of the infections caused by extensively drug-resistant Gram-negative bacilli: a Chinese consensus statement. Clin Microbiol Infect. 2016;22 Suppl 1(suppl 1):S15-25S15: S1. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



