Back to Journals » Advances and Applications in Bioinformatics and Chemistry » Volume 14
An Overview of Current Uses and Future Opportunities for Computer-Assisted Design of Vaccines for Neglected Tropical Diseases
Authors Robleda-Castillo R, Ros-Lucas A , Martinez-Peinado N, Alonso-Padilla J
Received 20 October 2020
Accepted for publication 3 January 2021
Published 15 February 2021 Volume 2021:14 Pages 25—47
DOI https://doi.org/10.2147/AABC.S258759
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Juan Fernandez-Recio
Raquel Robleda-Castillo, Albert Ros-Lucas, Nieves Martinez-Peinado, Julio Alonso-Padilla
Barcelona Institute for Global Health (ISGlobal), Hospital Clínic – University of Barcelona, Barcelona, 08036, Spain
Correspondence: Julio Alonso-Padilla Tel +34-932275400 (Ext. 4569)
Email [email protected]
Abstract: Neglected tropical diseases are infectious diseases that impose high morbidity and mortality rates over 1.5 billion people worldwide. Originally restricted to tropical and subtropical regions, changing climate conditions have increased their potential to emerge elsewhere. Control of their impact suffers from shortages like poor epidemiological surveillance or irregular drug distribution, and some NTDs still lack of appropriate diagnostics and/or efficient therapeutics. For these, availability of vaccines to prevent new infections, or the worsening of those already established, would mean a major breakthrough. However, only dengue and rabies count with approved vaccines at present. Herein, we review the state-of-the-art of vaccination strategies for NTDs, setting the focus on third generation vaccines and the concept of reverse vaccinology. Its capability to address pathogens´ biological complexity, likely contributing to save developmental costs is discussed. The use of computational tools is a fundamental aid to analyze increasingly large datasets aimed at designing vaccine candidates with the highest, possibly, opportunities to succeed. Ultimately, we identify and analyze those studies that took an in silico approach to find vaccine candidates, and experimentally assessed their immunogenicity and/or protection capabilities.
Keywords: neglected tropical diseases, vaccines, reverse vaccinology, immunoinformatics, epitopes
Introduction
Neglected Tropical Diseases (NTDs) are high-morbidity, high-mortality infectious diseases associated with extreme poverty in tropical and sub-tropical countries, where people usually have limited access to health services.1,2 They can be caused by bacteria, endo- and ecto-parasites, viruses and fungi. More recently, snake envenoming has also been included in the World Health Organization (WHO) list of NTDs.3 At present, this list includes 20 diseases (Table 1), which altogether are estimated to affect over 1.5 billion people worldwide, standing as a major obstacle to health access, poverty reduction, and socio-economic development.1–3
 |
Table 1 WHO List of NTDs and Their Corresponding Etiological Agents |
Most NTDs are zoonotic infections, but there are also others whose main reservoir are humans like dengue.2 Some are transmitted by vectors, while others spread by direct contact and/or oral and food-borne routes.2 Originally confined to tropical and sub-tropical regions, under currently changing climate conditions, their potential to emerge in other areas due to migration of human populations and/or changes in vector distribution must be taken into consideration. This is what respectively occurred with Chagas disease and Chikungunya virus in the last few decades.4,5 Therefore, it is very important to invest in them, as human health is a global matter. Increasing our comprehension and knowledge of NTDs to pursue their cure, epidemiological control and eventual eradication should be a priority to reduce their economic and social impact as well as to limit their chances of expansion.
Effective control of NTDs impact depends on access to appropriate diagnostics and the availability of efficient and safe drugs.6 In these regards, a “Preventive Chemotherapy and Transmission control (PCT)” approach is used on the group of NTDs that already have existing tools for their prevention and treatment, such as lymphatic filariasis, onchocerciasis or schistosomiasis.3,7 Nonetheless, success of PCT depends on the ability to identify populations at higher risk of suffering the disease impact and treat them, as well as controlling the causative vectors or any other possibilities of transmission. By contrast, there are other NTDs for which the implementation of appropriate tools for their detection and management is still lacking. For these, the strategy followed is the “Innovative and Intensified Disease Management (IDM)”,3,8 but research investment is low, their burden is not fully known, and it is urgent to improve already available interventions to control them, as well as identifying and developing novel ones.8 Several pharmaceutical companies, non-profit organizations, and governmental and academic agencies have provided economic aid to achieve this goal,9 and hopefully all those efforts will not wane.
For controlling disease impact, vaccinology is a field with great opportunity. Contribution of vaccines towards societal development by improvement of health status and increasing life-expectancy has been paramount. Many communicable diseases that used to exert a very high toll have been controlled or even eradicated (like smallpox) thanks to implementation of generalized vaccination programs.
Initially, vaccines were made by growing the pathogen in question and injecting a killed or attenuated version of it to evoke a protective memory immune response, in what has been termed “vaccinology 1.0.”10 With the advent of recombinant DNA technologies, new vaccination approaches have been explored immunizing with pathogen subunits or chimeric moieties, which led to “vaccinology 2.0.”10 Nowadays, advances in genetic sequencing and omics sciences allow a tailored design of vaccine candidates based on the genetic information of the pathogen,10 which coined the term “reverse vaccinology.”11 This methodology has already proved successful in the development of a vaccine against the challenging bacterial meningitis caused by meningococcus serogroup B (MenB) (trade name Bexsero).10,11 Such an approach is also known as “vaccinology 3.0,”10 and inspired by Bexsero, several vaccine candidates against other infectious diseases and cancer are being pursued following such approach.12–14
In that context, access to genomic and proteomic information of the pathogen is essential, and for the analysis and query of large genomic databases, a series of bioinformatic resources have been developed in the last few years (see Immunoinformatics Pipeline).11 However, despite the clear biomedical advantages offered by vaccination over the highly toxic chemotherapies employed to treat NTDs, their vaccine landscape remains mostly unpopulated. The complex biology of the microorganisms responsible for these diseases (many of them eukaryotic protozoa or even multicellular pathogens), and the very scarce funding dedicated to their research, have precluded vaccine development. With NTDs out of the scope of pharmaceutical industry portfolios, academic laboratories involved in the study of the biology and clinical outcome of these diseases stand as the main players in the field.
The aim of the present work was to review the state-of-the-art of vaccine development for NTDs, with a focus on technologically advanced third generation vaccines. These could represent the sought after solution to ameliorate their impact. We framed the information retrieved on the availability of genomic data of the pathogens, and systematically identified those studies that have actually surpassed the initial computational part of the work and included experimental data on immunogenicity and protection against infection.
Search Strategy and Eligibility Criteria
We undertook the search of publications in PubMed/MEDLINE database. We included publications on clinical studies, clinical trials, reviews, and journal articles that assessed NTDs in the WHO list and/or their causative agents, according to the search terms depicted in Table 2. Searches were performed in March 2020, and we restricted the publications retrieved to those published within the last 20 years. Figure 1 depicts the number of publications found per NTD or pathogen according to the search terms used. We performed a manual revision to discard any non-relevant publications, while full texts presenting experimental data were individually curated to select those that included immunogenicity and/or protection data of the vaccine candidates under research. Biological information available for each pathogen was retrieved at the corresponding NCBI repositories. Identification of the number of available genomes for each of the NTDs etiological agents was carried out in the NCBI Genome data base.
 |
Table 2 Keywords Used in the Electronic Searches |
The Vaccine Landscape for NTDs: Where are We?
Some NTDs already have efficient chemotherapies at hand while others do not. Although these are not absent of limitations, like short supply and lack of distribution, Buruli ulcer, foodborne trematodiases, leprosy, onchocerciasis, scabies, schistosomiasis, soil-transmitted helminthiases, snakebite envenoming, taeniasis/cysticercosis, trachoma and yaws belong to a group of NTDs that generally count with effective therapeutics available. On the contrary, a second group of diseases encompassing Chagas disease, dengue, chikungunya, dracunculiasis, echinococcosis, human (and animal) African trypanosomiasis, leishmaniasis, lymphatic filariasis and rabies, could largely benefit from improved therapeutic solutions. For the treatment of some of the latter (like Chagas disease, African trypanosomiasis, leishmaniasis and lymphatic filariasis) there are drugs available, but they are either too expensive, entail invasive administration, render variable efficacies, and/or have frequent side-effects. For others, mainly viral infections like dengue, chikungunya or rabies, but also dracunculiasis, there are no drugs at all. Mycetoma and chromoblastomycosis would fit in both groups, depending on their causative agent: if it is bacteria, treatments are usually secure and effective; if it is fungi, available therapies are more expensive and not exempt from numerous adverse effects. Anyhow, for all those NTDs that lack of an appropriate and accessible treatment, vaccination represents a very valuable solution. In fact, two of those NTDs, dengue and rabies, currently have approved vaccines to avert them.
Dengue and Rabies Vaccines
Dengue vaccine was developed by Sanofi Pasteur, which commercializes it under the trade name Dengvaxia® (CYD-TVD).15 It is based on live attenuated dengue viruses and was approved in 2015 for vaccinating people between nine and 45 years old.15 CYD-TVD is a tetravalent vaccine that targets the four existing serotypes of the virus causing dengue disease (DENV-1, DENV-2, DENV-3, DENV-4), but yields serotype-variable efficiencies.15 In fact, efficacy against the highly virulent serotype 2 was found to be the lowest of the four.15 Its induced immunogenicity depends on several factors such as age, immune status of the patient, other underlying diseases, and/or previous exposure to the virus.15 In relation to the latter, a recent study reported it was less efficient in seronegative people, and worryingly, in some cases it was found that its use could entail severe forms of dengue upon occurrence of posterior infections.15,16
In the case of rabies, there are several vaccines commercialized. The first rabies vaccine was developed by Louis Pasteur in 1885 and consisted of inactivated viruses extracted from spinal cords of rabbits (Pasteur vaccine).17 It had safety risks due to the possible presence of live viruses, and for this reason the Semple rabies vaccine was developed adding phenol to completely inactivate them.17 Either way, both vaccines caused severe side effects due to the presence of myelin from the rabbits´ nervous system that was the source of the viruses.17 A third attempt to have a safer rabies vaccine was that of Fuenlazida product, obtained from newborn or embryonic mice, it contained less quantity of myelin.17 Nonetheless, it was not free from side-effects because there was still some presence of myelin in it.17 More recently, rabies vaccines have been obtained from infected cell cultures, which yielded products with increased immunogenicity and fewer adverse effects, so vaccine production is presently made in duck or chicken embryonic cells.17 All licensed options are based on inactivated viruses because the administration of attenuated live rabies virus to humans is not approved due to the possibility of developing the disease.18 Rabies vaccines like RabAvert, Rabipur or Rabivax efficiently induce long-lasting immunity triggering the production of neutralizing antibodies but several doses are required.18 Regrettably, rabies vaccination is not included in the vaccines covered by the Expanded Programme on Immunization, and they are expensive and not affordable in developing countries.19 Furthermore, real demand is difficult to calculate, which leads to shortage of doses and purchasing of poor-quality vaccine alternatives at higher costs and without quality assurance.20 Thus, investigations are ongoing to find cost-effective and efficient alternatives, but most new vaccine candidates have not been clinically tested yet.18
Next Generation Vaccinology for NTDs
Most NTDs are caused by biologically complex eukaryotic parasites or even multicellular organisms, and current vaccination approaches may not be sufficient to tackle such complexity. Fortunately, vaccinology is a dynamically evolving research area and new vaccine development strategies such as reverse vaccinology are now available. Principles of vaccinology 1.0 and 2.0 led to the development of numerous vaccine products for several infectious diseases, but their whole undertaking is time consuming, arduous and very expensive.10 Moreover, they have been unable to confer protection against numerous emerging and re-emerging infections.21 Therefore, thinking of NTDs context of high medical urgency and low investment, new strategies involving shorter development times and lower costs would be required.
In the last few decades, linked to the latest advances in sequencing and omics sciences, we have witnessed the expansion and perfecting of a plethora of in silico tools, which application could contribute to the design of vaccine candidates at a fraction of the cost of more traditional methods. Experimentally screening large genomes to find out the antigens of vaccine interest would be highly time-consuming and costly.22 In this regards, implementation of vaccinology 3.0 and its related immunoinformatics methodologies to in silico pre-screening of the pathogens genomes would guide the selection of those antigens with higher interest.22 Such tools are also very valuable if the pathogen in question is isolated and grown with difficulty. In addition, in silico-based approaches may permit seeking “pan-genome” protective antigens, exploring vaccine candidates that can immunize against the different pathogenic strains or genetic variants of the pathogen.22–24 Another advantage of identifying evolutionary conserved sequences would be that conservation might respond to the importance of such sequences in the pathogen life cycle, making of them remarkable vaccine targets. Thus, combination of in silico methods with omics to prioritize the most suitable sequences to be specifically investigated experimentally, in vitro and in vivo, stands as a promising approach to develop new vaccines. Nowadays there are several efforts following these principles to design vaccines for NTDs, their number varying depending on the research background and specificities of each disease (Table 3). The strategy has also been used to investigate vaccine candidates against protozoan parasites like Toxoplasma gondii and Neospora caninum.25,26
 |  |  |  |  |  |
Table 3 Studies Following a Reverse Vaccinology Approach to Develop Vaccine Candidates for NTDs |
When correlating NTDs vaccinology research with the number of published studies, we can see that the commitment is very unequal depending on the disease being considered. Some seem to receive little to no interest when it comes to the development of a vaccine or vaccine-like approach, like Buruli ulcer, trachoma or Yaws, because understandably they already dispose of treatments against them (Figure 1B). With respect to the retrieved studies on NTDs vaccinology 3.0 that included wet lab experimental results, most have been performed in mice and rabbits as pre-clinical models of infection, with the exception of a study performed on non-human primates (Table 3). Other works relied on patients’ peripheral blood mononuclear cells (PBMCs) to run epitope identification experiments. When it comes to the epitopes selected to perform the immunizations, both T cell epitopes and B cell epitopes were used, which in the majority of the cases were administered in the presence of an adjuvant (Table 3). Techniques to perform the analysis of immune responses by either in vivo infection models or samples collected from patients included enzyme-linked immunosorbent assays (ELISAs), intracellular cytokine staining and ELISPOTs.
In connection with the higher funding and interest received, dengue is the disease with more studies exploring a reverse vaccinology approach (Table 3). Most of them are based on the use of peptides from the envelope (E) glycoprotein as immunogens (Table 3). This protein constitutes the primary target for inducing neutralizing antibodies since it interacts with receptors of host cells to mediate virus internalization. Moreover, it triggers a T cell response after virus infection that plays a vital role in providing protection against it. A few other studies are based on targeting non-structural proteins (NS) which are highly immunogenic. Commonly, all report some sort of protection and immunological response, but in those where different serotypes were studied, immunogenicity and protection was not induced to all of them, highlighting the relevance of selecting invariable regions to call the immunity (Table 3).
Publications on leishmaniasis are the second most numerous overall (Figure 1). When it comes to vaccinology 3.0 efforts, most are in vitro works using patients´ PBMCs stimulated with a variety of antigens. A remarkable one is thiol-specific antioxidant (TSA), a promising vaccine candidate due to its protective capacity against different Leishmania spp. (Table 3).
Studies on vaccines for schistosomiasis are mostly based on Sm200, a tegument protein, and different secretory and transmembrane proteins. While Sm200 protein was unable to reduce worm or egg burdens, secretory and transmembrane proteins increased T-cell proliferation and production of interferon gamma (Table 3). These results make of the selected epitopes within them promising candidates for a possible future epitope-based vaccine.
Echinococcosis is another NTD endowed with work on vaccinology as a therapeutic approach. Eg95 antigen is the common molecule used in the two available studies (Table 3). Thought to influence host specificity and affect vaccine efficiency due to its increased expression on the parasite during invasion of host cells, results revealed the antigenic capability of this antigen. When combined with other proteins of the parasite in a chimeric molecule, it led to an increased protection and a reduced number of cysts when compared to a control group in the in vivo models of infection (Table 3).
Studies on Chagas disease were also identified, mostly based on immunization with peptides obtained from MASP family members and trans-sialidases (Table 3). Proteins from these families are essential for a successful infection as they play an important role in the host-parasite interaction and in the establishment of the disease chronic stage.40 In vivo studies showed some kind of immunization in mice, increasing their survival rate, production of antibodies and managed to decrease the parasitic load (Table 3).
Finally, a single study was found related to the development of a vaccine for onchocerciasis (Table 3). It was performed using PBMCs isolated from patients and led to the discovery of promising peptides for a possible diagnosis of the disease, as there is evident recognition on the part of the infected patients’ cells. For obtaining an immunization output, though, further investigations will be required.
Aforementioned diseases with studies that bestow real data on the development of in silico designed epitope-based vaccines are characterized by the fact that they lack efficient and safe treatments, and a real solution is yet to be discovered. In comparison to dengue, Chagas disease, echinococcosis, leishmaniasis, onchocerciasis and schistosomiasis are caused by parasites with much larger genomes. A feature reflected in the wide variability of available immunogens for each parasite that results in an increased difficulty to ultimately develop a vaccine for them. Their genomes have been sequenced (eg, for T. cruzi there are several strains already sequenced), and there are several immunoinformatic tools that can aid in the selection of those antigens or epitopes of interest to be further studied for their inclusion in potential vaccine candidates. Notably, there are improved in vivo models of disease where those can be evaluated, in some cases even including non-human primates. Plus the analysis of in silico identified epitopes can also be made with clinical samples from patients and controls, which may contribute to a rapid translation of results reducing the attrition rates.
Immunoinformatics Pipeline
The largest publicly available repository of epitopes is the Immune Epitope Database (IEDB), funded by the United States of America National Institutes of Health (NIH).52 This open resource holds data on more than 900,000 epitopes, products of the curation of immunologic assays in the scientific literature, and includes their reactivity, type and grade of response, antigens of origin, among information of relevance.53 In addition, one of the available options when querying the database is the selection of those epitopes that have been validated experimentally. It also features the IEDB Analysis Resource, a hub of immunoinformatics tools for epitope prediction and assessment.
In search of potentially good epitopes to encompass a vaccine ensemble, protein invariability has been considered to be a major attribute.23,24,54,55 A hypothetical vaccine product providing pan-protection against multiple strains of a circulating pathogen would indeed be a much desirable characteristic considering the very limited resources for vaccine development, especially for NTDs. Therefore, an initial step to find out the best epitopes often entails using CD-HIT or other BLAST-like tools to cluster together all the proteins from the different proteomes or strains of the pathogen in question, in order to reduce their redundancy (Figure 2).
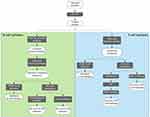 |
Figure 2 Flowchart depicting the procedures followed to reach selected B-cell and T-cell epitopes. In each case, the legacy experimentation approach is indicated on the left. |
Each of these protein clusters will then undergo a multiple sequence alignment step (eg, using MUSCLE), so that the variability within each amino acid position can be further analyzed. A measurement that is applied to parametrize the variability is the Shannon entropy,56 which quantifies it and allows masking of any position with variability over a certain entropy threshold.57 The outcome will be a conserved consensus proteome of our pathogen of interest, against which the list of experimentally validated epitopes retrieved from the IEDB repository can be parsed following a legacy experimentation approach.58 However, basic research on NTDs and their etiological pathogens has historically been neglected, and the amount of information on their epitopes is usually insufficient. As a consequence, a legacy experimentation approach cannot be pursued. Fortunately, there is a whole series of tools designed and trained for de novo prediction of epitopes.
Adaptive immunity main players are T cells and B cells, so epitopes are consequently classified as T-cell or B-cell epitopes. The former can be further differentiated into CD8+ or CD4+ T-cell epitopes, the two main subpopulations of these cells. Predictions for T-cell epitopes mainly focus on MHC binding.59 The IEDB Analysis Resource hosts several prediction tools for both types of T-cell epitopes, with a default “IEDB recommended” method that is periodically updated based on benchmarks of predictors for a specific allele.60 Generally speaking, binding prediction tools for MHC I (CD8+) molecules are more accurate than for MHC II (CD4+).59 Additionally, MHC I predictions can be somewhat enhanced by taking into account intracellular antigen processing steps, such as proteasomal degradation and the importation of potential epitopes into the endoplasmic reticulum through TAP, the transporter associated with antigen processing.61 This allows the creation of multi step prediction methods for CD8+ T-cell epitopes, which can be combined into a single tool (http://tools.iedb.org/processing/), or used separately.62,63
On the other hand, B-cell epitope predictions are more unreliable than those of T-cell epitopes.59 B-cell epitopes drive humoral immune responses based on antibodies. Some are conformational (discontinuous) epitopes, while others are linear (continuous). Most tools are focused on predicting the latter because they can be directly predicted out of the protein primary structure and delivered independently from their context in order to induce an immune response, while that is much more complex to achieve with discontinuous epitopes.59 An alternative approach for B-cell epitopes prediction is to rely on 3D protein structures where it is possible to identify regions accessible to antibody recognition through relative solvent accessibility calculations (eg, using NACCESS64), as well as determining the flexibility of such regions by analyzing the temperature factors, or B-factors, of residues within 3D structures.23,24 These factors are included in .pdb files downloaded from the Protein Data Bank,65 and are a measure of the smearing in a protein structure due to the motions of its atoms: the higher this value is, the higher the flexibility.66
Predicted epitopes can then be packaged together to construct a new vaccine ensemble. However, when designing a new epitope-based vaccine, two main issues have to be taken into account: which combination of epitopes will offer the highest immune coverage, and how are these administered.67,68 The former can be answered using HLA coverage tools, which output is the best combination of epitopes for a wider coverage taking into consideration the population frequencies of HLA alleles. Two advantages of prioritizing epitopes following this approach are envisaged: first, ensuring selected epitopes are recognized by HLA alleles with a high population frequency,68 and second, targeting specific HLAs considering different frequencies for different human populations.68 Examples of these tools include the Population Coverage tool at the IEDB Analysis Resource,53 PEPVAC,69 and PREDIVAC.70 Regarding the delivery of the epitope-based vaccine candidates, poor immunogenicity has been described when administering them as peptides,54 so the use of genetic delivery has also been proposed.23,24 In any case, the use of vaccine encoded adjuvant sequences or the co-administration of the vaccine constructs with an adjuvant would be desired. Finally, selected epitopes have to be delivered in a vaccine formulation, usually in the form of a multi-epitope “string of beads” genetic construct.67 Any theoretical construct must take into account the order of the epitopes, as well as the separation between them, in order to avoid the appearance of neo-epitopes, which could have negative effects.71 The most common approach is to use “linker” or “spacer” sequences that promote proteasomal cleavage, such as the commonly used AAY sequence.67,71 Additionally, some algorithms have been developed to facilitate this task, such as the one developed by Schubert and Kohlbacher,71 or VaccineCAD.72
All these tools could play a critical role in the search for vaccines against NTDs due to their effects in the reduction of costs. “Traditional” epitope identification involves costly time-consuming screenings of large arrays of candidates, and being able to reduce the potential list of epitopes via the methodologies described above might provide a huge benefit.
Conclusions
No doubt implementing vaccination programs would be a breakthrough for controlling the impact of several NTDs. Nonetheless, this has been generally poorly explored due to scarcity of funding resources and the biological complexity of the pathogens causing them. Nowadays, new vaccinology approaches are at hand opening a field of great possibilities for the cost-effective design and assessment of vaccine candidates against complex pathogens carrying thousands of potential antigens. Yet, investments will be needed to progress on this complicated challenge. To this day, dengue and rabies are the only NTDs that can be vaccine prevented, but issues like cost and availability still limit their more widespread use.
Ongoing research efforts onto NTDs vaccines are unevenly distributed. If it is true that some NTDs would not require a vaccine due to the current existence of efficacious treatments, others could greatly benefit from them. For that, a computer-assisted strategy to screen the pathogens´ extensive antigen repertoire could largely contribute to save developmental costs. This might also be applied to the design of epitope-based vaccine candidates which could be at the same time a major approach to counteract the high biological complexity of the pathogens by allowing inclusion of multiple epitopes from multiple antigens.
Either way, progress towards NTDs’ control and elimination mostly rest on academic groups under the umbrella of international partnerships funds and donations from pharmaceutical companies, but there is still a long road ahead. Strong and continued commitment by all actors involved in this process will be fundamental to eventually succeed in the control of NTDs impact.
Consent for Publication
We give our consent for the publication of this paper and its contents.
Acknowledgments
We are thankful to Drs Joaquim Gascón and Maria-Jesus Pinazo from ISGlobal Chagas Initiative for their continuous support and intellectual feedback.
Author Contributions
All authors made a significant contribution to the work reported, whether that was in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
We want to thank the support by the Departament d’Universitats i Recerca de la Generalitat de Catalunya, Spain (AGAUR; 2017SGR00924), and the funding by the Instituto de Salud Carlos III RICET Network for Cooperative Research in Tropical Diseases (ISCIII; RD12/0018/0010) and FEDER. We also acknowledge support from the Spanish Ministry of Science, Innovation and Universities through the “Centro de Excelencia Severo Ochoa 2019–2023” Program (CEX2018-000806-S), and support from the Generalitat de Catalunya through the “CERCA Program.”
Disclosure
The authors report no conflicts of interest in this work.
References
1. Molyneux DH, Savioli L, Engels D. Neglected tropical diseases: progress towards addressing the chronic pandemic. Lancet. 2017;389(10066):312–325. doi:10.1016/S0140-6736(16)30171-4
2. Klohe K, Amuasi J, Kaducu JM, et al. The 2017 Oslo conference report on neglected tropical diseases and emerging/re-emerging infectious diseases - focus on populations underserved. Infect Dis Poverty. 2019;8(1):40. doi:10.1186/s40249-019-0550-8
3. WHO. World Health Organization. Available from: https://www.who.int/neglected_diseases/diseases/en/.
4. Pérez-Molina JA, Molina I. Chagas disease. Lancet. 2018;391(10115):82–94. doi:10.1016/S0140-6736(17)31612-4
5. Silva LA, Dermody TS. Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J Clin Invest. 2017;127(3):737–749. doi:10.1172/JCI84417
6. Rosenberg M, Utzinger J, Addiss DG, Hirayama K. Preventive chemotherapy versus innovative and intensified disease management in neglected tropical diseases: a distinction whose shelf life has expired. PLoS Negl Trop Dis. 2016;10(4):e0004521. doi:10.1371/journal.pntd.0004521
7. Akinsolu FT, Nemieboka PO, Njuguna DW, Ahadji MN, Dezso D, Varga O. Emerging resistance of neglected tropical diseases: a scoping review of the literature. Int J Environ Res Public Health. 2019;16(11):1925. doi:10.3390/ijerph16111925
8. WHO. Innovative and intensified disease management (IDM). Available from: https://www.who.int/neglected_diseases/disease_management/Innovative_Intensified_Disease_Management/en/.
9. Nauleau P, Ralph H. Neglected tropical diseases: research bites back. EBioMedicine. 2016;11:1. doi:10.1016/j.ebiom.2016.09.009
10. Bragazzi NL, Gianfredi V, Villarini M, et al. Vaccines meet big data: state-of-the-art and future prospects. From the classical 3Is (“Isolate-Inactivate-Inject”) vaccinology 1.0 to vaccinology 3.0, vaccinomics, and beyond: a historical overview. Front Public Health. 2018;6:62. doi:10.3389/fpubh.2018.00062
11. Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33(4):530–541. doi:10.1016/j.immuni.2010.09.017
12. de Sarom A, Kumar Jaiswal A, Tiwari S, et al. Putative vaccine candidates and drug targets identified by reverse vaccinology and subtractive genomics approaches to control Haemophilus ducreyi, the causative agent of chancroid. J R Soc Interface. 2018;15(142):20180032. doi:10.1098/rsif.2018.0032
13. Capelli R, Peri C, Villa R, et al. BPSL1626: reverse and structural vaccinology reveal a novel candidate for vaccine design against Burkholderia pseudomallei. Antibodies. 2018;7(3):26. doi:10.3390/antib7030026
14. Wang X, Liu Y, Diao Y, et al. Gastric cancer vaccines synthesized using a TLR7 agonist and their synergistic antitumor effects with 5-fluorouracil. J Transl Med. 2018;16(1):120. doi:10.1186/s12967-018-1501-z
15. Malisheni M, Khaiboullina SF, Rizvanov AA, Takah N, Murewanhema G, Bates M. Clinical efficacy, safety, and immunogenicity of a live attenuated tetravalent dengue vaccine (CYD-TDV) in children: a systematic review with meta-analysis. Front Immunol. 2017;8:863. doi:10.3389/fimmu.2017.00863
16. Fatima K, Syed NI. Dengvaxia controversy: impact on vaccine hesitancy. J Glob Health. 2018;8(2):020312. doi:10.7189/jogh.08.020312
17. Zhu S, Guo C. Rabies control and treatment: from prophylaxis to strategies with curative potential. Viruses. 2016;8(11):279. doi:10.3390/v8110279
18. Fisher CR, Schnell MJ. New developments in rabies vaccination. Rev Sci Tech. 2018;37(2):657–672. doi:10.20506/rst.37.2.2831
19. WHO. The expanded programme on immunization. Available from: https://www.who.int/immunization/programmes_systems/supply_chain/benefits_of_immunization/en/.
20. Abela-Ridder B, Martin S, Gongal G, Engels D. Rabies vaccine stockpile: fixing the supply chain. Bull World Health Organ. 2016;94(9):635–635A. doi:10.2471/BLT.16.183012
21. Pritam M, Singh G, Swaroop S, Singh AK, Singh SP. Exploitation of reverse vaccinology and immunoinformatics as promising platform for genome-wide screening of new effective vaccine candidates against Plasmodium falciparum. BMC Bioinform. 2019;19(13):468. doi:10.1186/s12859-018-2482-x
22. Naz K, Naz A, Ashraf ST, et al. PanRV: pangenome-reverse vaccinology approach for identifications of potential vaccine candidates in microbial pangenome. BMC Bioinform. 2019;20(1):123. doi:10.1186/s12859-019-2713-9
23. Michel-Todó L, Reche PA, Bigey P, Pinazo MJ, Gascón J, Alonso-Padilla J. In silico design of an epitope-based vaccine ensemble for chagas disease. Front Immunol. 2019;10:2698. doi:10.3389/fimmu.2019.02698
24. Michel-Todó L, Bigey P, Reche PA, Pinazo MJ, Gascón J, Alonso-Padilla J. Design of an epitope-based vaccine ensemble for animal trypanosomiasis by computational methods. Vaccines. 2020;8(1):130. doi:10.3390/vaccines8010130
25. Goodswen SJ, Kennedy PJ, Ellis JT. A gene-based positive selection detection approach to identify vaccine candidates using Toxoplasma gondii as a test case protozoan pathogen. Front Genet. 2018;9:332. doi:10.3389/fgene.2018.00332
26. Goodswen SJ, Kennedy PJ, Ellis JT. On the application of reverse vaccinology to parasitic diseases: a perspective on feature selection and ranking of vaccine candidates. Int J Parasitol. 2017;47(12):779–790. doi:10.1016/j.ijpara.2017.08.004
27. Pinto PBA, Assis ML, Vallochi AL, et al. T cell responses induced by DNA vaccines based on the DENV2 E and NS1 proteins in mice: importance in protection and immunodominant epitope identification. Front Immunol. 2019;10:1522. doi:10.3389/fimmu.2019.01522
28. Huang X, Karabudak A, Comber JD, Philip M, Morcol T, Philip R. A novel immunization approach for dengue infection based on conserved T cell epitopes formulated in calcium phosphate nanoparticles. Hum Vaccin Immunother. 2017;13(11):2612–2625. doi:10.1080/21645515.2017.1369639
29. Ramasamy V, Arora U, Shukla R, et al. A tetravalent virus-like particle vaccine designed to display domain III of dengue envelope proteins induces multi-serotype neutralizing antibodies in mice and macaques which confer protection against antibody dependent enhancement in AG129 mice. PLoS Negl Trop Dis. 2018;12(1):e0006191. doi:10.1371/journal.pntd.0006191
30. Chen HW, Hu HM, Wu SH, et al. The immunodominance change and protection of CD4+ T-cell responses elicited by an envelope protein domain III-based tetravalent dengue vaccine in mice. PLoS One. 2015;10(12):e0145717. doi:10.1371/journal.pone.0145717
31. Dos Santos Alves RP, Pereira LR, Fabris DLN. Production of a recombinant dengue virus 2 NS5 protein and potential use as a vaccine antigen. Clin Vaccine Immunol. 2016;23(6):460–469. doi:10.1128/CVI.00081-16
32. Quinan BR, Flesch IEA, Pinho TMG, Coelho FM, Tscharke DC, da Fonseca FG. An intact signal peptide on dengue virus E protein enhances immunogenicity for CD8+ T cells and antibody when expressed from modified vaccinia Ankara. Vaccine. 2014;32(25):2972–2979. doi:10.1016/j.vaccine.2014.03.093
33. Chen XY, Li DZ, Zhong XZ, Chen B, Duan ZL, Wen JS. Induction of multiple cytotoxic T lymphocyte responses in mice by a multiepitope DNA vaccine against dengue virus serotype 1. Microbiol Immunol. 2016;60(12):835–845. doi:10.1111/1348-0421.12457
34. Roth C, Cantaert T, Colas C, et al. A modified mRNA vaccine targeting immunodominant NS epitopes protects against dengue virus infection in HLA class I transgenic mice. Front Immunol. 2019;10:1424. doi:10.3389/fimmu.2019.01424
35. Tripathi L, Mani S, Raut R, et al. Pichia pastoris-expressed dengue 3 envelope-based virus-like particles elicit predominantly domain III-focused high titer neutralizing antibodies. Front Microbiol. 2015;6:1005. doi:10.3389/fmicb.2015.01005
36. Nascimento EJM, Mailliard RB, Khan AM, et al. Identification of conserved and HLA promiscuous DENV3 T-cell epitopes. PLoS Negl Trop Dis. 2013;7(10):e2497. doi:10.1371/journal.pntd.0002497
37. da Silva ANMR, Nascimento EJM, Cordeiro MT, et al. Identification of continuous human B-cell epitopes in the envelope glycoprotein of dengue virus type 3 (DENV-3). PLoS One. 2009;4(10):e7425. doi:10.1371/annotation/238cbff8-6794-4796-8a8a-a80d3b246757
38. Ramanathan B, Poh CL, Kirk K, McBride WJH, Aaskov J, Grollo L. Synthetic B-cell epitopes eliciting cross-neutralizing antibodies: strategies for future dengue vaccine. PLoS One. 2016;11(5):e0155900. doi:10.1371/journal.pone.0155900
39. Khetarpal N, Shukla R, Rajpoot RK, et al. Recombinant dengue virus 4 envelope glycoprotein virus-like particles derived from Pichia pastoris are capable of eliciting homotypic domain III-directed neutralizing antibodies. Am J Trop Med Hyg. 2017;96(1):126–134. doi:10.4269/ajtmh.16-0503
40. Serna C, Lara JA, Rodrigues SP, Marques AF, Almeida IC, Maldonado RA. A synthetic peptide from Trypanosoma cruzi mucin-like associated surface protein as candidate for a vaccine against Chagas disease. Vaccine. 2014;32(28):3525–3532. doi:10.1016/j.vaccine.2014.04.026
41. Alvarez MG, Postan M, Weatherly DB, et al. HLA class I-T cell epitopes from trans-sialidase proteins reveal functionally distinct subsets of CD8+ T cells in chronic Chagas disease. PLoS Negl Trop Dis. 2008;2(9):e288. doi:10.1371/journal.pntd.0000288
42. Wang L, Gao J, Lan X, et al. Identification of combined T-cell and B-cell reactive Echinococcus granulosus 95 antigens for the potential development of a multi-epitope vaccine. Ann Transl Med. 2019;7(22):652. doi:10.21037/atm.2019.10.87
43. Esmaelizad M, Ahmadian G, Aghaiypour K, Shamsara M, Paykari H, Tebianian M. Induction of protective T-helper 1 immune responses against Echinococcus granulosus in mice by a multi-T-cell epitope antigen based on five proteins. Mem Inst Oswaldo Cruz. 2013;108(4):408–413. doi:10.1590/S0074-0276108042013003
44. Pandey RK, Dikhit MR, Kumar A, et al. Evaluating the immunomodulatory responses of LdODC-derived MHC Class-II restricted peptides against VL. Parasite Immunol. 2020;42(4):e12699. doi:10.1111/pim.12699
45. Dikhit MR, Das S, Mahantesh V, et al. The potential HLA Class I-restricted epitopes derived from LeIF and TSA of Leishmania donovani evoke anti-leishmania CD8+ T lymphocyte response. Sci Rep. 2018;8(1):14175. doi:10.1038/s41598-018-32040-x
46. Dikhit MR, Kumar A, Das S, et al. Identification of potential MHC Class-II-restricted epitopes derived from Leishmania donovani antigens by reverse vaccinology and evaluation of their CD4+ T-cell responsiveness against visceral leishmaniasis. Front Immunol. 2017;8:1763. doi:10.3389/fimmu.2017.01763
47. De Brito RCF, De Oliveira Cardoso JM, Reis LES, et al. Synthetic peptides elicit strong cellular immunity in visceral leishmaniasis natural reservoir and contribute to long-lasting polyfunctional T-cells in BALB/c mice. Vaccines. 2019;7(4):162. doi:10.3390/vaccines7040162
48. Lagatie O, Verheyen A, Van Dorst B, Batsa Debrah L, Debrah A, Stuyver LJ. Linear epitopes in Onchocerca volvulus vaccine candidate proteins and excretory-secretory proteins. Parasite Immunol. 2018;40(11):e12587. doi:10.1111/pim.12587
49. Carvalho GBF, De, Pacífico LGG, Pimenta DLF. Evaluation of the use of C-terminal part of the Schistosoma mansoni 200kDa tegumental protein in schistosomiasis diagnosis and vaccine formulation. Exp Parasitol. 2014;139(1):24–32. doi:10.1016/j.exppara.2014.02.003
50. de Souza C, Lopes MD, De Oliveira FM, et al. Rational selection of immunodominant and preserved epitope Sm043300e from Schistosoma mansoni and design of a chimeric molecule for biotechnological purposes. Mol Immunol. 2018;93:133–143. doi:10.1016/j.molimm.2017.11.019
51. Zhao BP, Chen L, Zhang YL, et al. In silico prediction of binding of promiscuous peptides to multiple MHC class-II molecules identifies the Th1 cell epitopes from secreted and transmembrane proteins of Schistosoma japonicum in BALB/c mice. Microbes Infect. 2011;13(7):709–719. doi:10.1016/j.micinf.2011.03.005
52. National Institutes of Health (NIH). Available from: https://www.nih.gov/.
53. Vita R, Mahajan S, Overton JA, et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–D343. doi:10.1093/nar/gky1006
54. Toussaint NC, Maman Y, Kohlbacher O, Louzoun Y. Universal peptide vaccines - optimal peptide vaccine design based on viral sequence conservation. Vaccine. 2011;29(47):8745–8753. doi:10.1016/j.vaccine.2011.07.132
55. Sheikh QM, Gatherer D, Reche PA, Flower DR. Towards the knowledge-based design of universal influenza epitope ensemble vaccines. Bioinformatics. 2016;32(21):3233–3239. doi:10.1093/bioinformatics/btw399
56. Shannon CEA. Mathematical theory of communication. Bell Syst Tech J. 1948;27(4):623–656. doi:10.1002/j.1538-7305.1948.tb00917.x
57. Stewart JJ, Lee CY, Ibrahim S, et al. A shannon entropy analysis of immunoglobulin and T cell receptor. Mol Immunol. 1997;34(15):1067–1082. doi:10.1016/S0161-5890(97)00130-2
58. Quinzo MJ, Lafuente EM, Zuluaga P, Flower DR, Reche PA. Computational assembly of a human cytomegalovirus vaccine upon experimental epitope legacy. BMC Bioinform. 2019;20(S6):476. doi:10.1186/s12859-019-3052-6
59. Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Fundamentals and methods for T- and B-cell epitope prediction. J Immunol Res. 2017;2017:2680160. doi:10.1155/2017/2680160
60. Dhanda SK, Mahajan S, Paul S, et al. IEDB-AR: immune epitope database - analysis resource in 2019. Nucleic Acids Res. 2019;47(W1):W502–W506. doi:10.1093/nar/gkz452
61. Doytchinova IA, Guan P, Flower DR. EpiJen: a server for multistep T cell epitope prediction. BMC Bioinform. 2006;7(1):131. doi:10.1186/1471-2105-7-131
62. Nielsen M, Lundegaard C, Lund O, Keşmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57(1–2):33–41. doi:10.1007/s00251-005-0781-7
63. Besser H, Louzoun Y. Cross-modality deep learning-based prediction of TAP binding and naturally processed peptide. Immunogenetics. 2018;70(7):419–428. doi:10.1007/s00251-018-1054-6
64. Hubbard SJ, Thornton JM. NACCESS, computer program. department of biochemistry and molecular biology, university college. London; 1993. Avaliable from: https://www.scirp.org/(S(351jmbntvnsjt1aadkposzje))/reference/ReferencesPapers.aspx?ReferenceID=68275.
65. Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi:10.1093/nar/28.1.235
66. PDB-101: learn: guide to understanding PDB data: dealing with coordinates. Available from: https://pdb101.rcsb.org/learn/guide-to-understanding-pdb-data/dealing-with-coordinates.
67. Terry FE, Moise L, Martin RF, et al. Time for T? Immunoinformatics addresses vaccine design for neglected tropical and emerging infectious diseases. Expert Rev Vaccines. 2014;14(1):21–35. doi:10.1586/14760584.2015.955478
68. Oli AN, Obialor WO, Ifeanyichukwu MO, et al. Immunoinformatics and vaccine development: an overview. ImmunoTargets Ther. 2020;9:13–30. doi:10.2147/ITT.S241064
69. Reche PA. Genome wide prediction of promiscuous epitope for vaccine design. Available from: http://imed.med.ucm.es/PEPVAC/.
70. Oyarzún P, Ellis JJ, Bodén M, Kobe B. PREDIVAC: CD4+ T-cell epitope prediction for vaccine design that covers 95% of HLA class II DR protein diversity. BMC Bioinform. 2013;14(1):52. doi:10.1186/1471-2105-14-52
71. Schubert B, Kohlbacher O. Designing string-of-beads vaccines with optimal spacers. Genome Med. 2016;8(1):9. doi:10.1186/s13073-016-0263-6
72. De Groot AS, Marcon L, Bishop EA, et al. HIV vaccine development by computer assisted design: the GAIA vaccine. Vaccine. 2005;23(17–18):2136–2148. doi:10.1016/j.vaccine.2005.01.097
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

