Back to Journals » Cancer Management and Research » Volume 10
An intronic genetic variation of MGMT affects enhancer activity and is associated with glioma susceptibility
Authors Huang L, Xu W, Dai L, Yan D, Zhang S, Shi X
Received 8 June 2018
Accepted for publication 6 August 2018
Published 27 September 2018 Volume 2018:10 Pages 3995—4003
DOI https://doi.org/10.2147/CMAR.S176622
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Liming Huang,1 Wenshen Xu,2 Lian Dai,3 Danfang Yan,4 Shu Zhang,1 Xi Shi1
1The First Department of Chemotherapy, The First Affiliated Hospital, Fujian Medical University, Fuzhou, Fujian, China; 2Department of Laboratory Medicine, The First Affiliated Hospital, Fujian Medical University, Fuzhou, Fujian, China; 3Department of Medicine, The Third Affiliated People’s Hospital, Fujian University of Traditional Chinese Medicine, Fuzhou, Fujian, China; 4Department of Radiation Oncology, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, Zhejiang, China
Purpose: O6-methylguanine-DNA methyltransferase (MGMT) plays a crucial role in repairing damaged DNA caused by alkylating agents. A number of cancer susceptibility loci have been recognized as enhancer variants. This study aimed to explore the significance of enhancer variants of MGMT in glioma susceptibility.
Patients and methods: A retrospective case-control study consisting of 150 glioma patients and 327 controls was conducted to test whether enhancer variants of MGMT are associated with glioma susceptibility. Genotypes were determined by Sequenom MassARRAY technology. Associations were estimated by logistic regression. Biochemical assays were used to examine the function of glioma susceptibility locus.
Results: We found that the A allele of rs10764901, an intronic variant of MGMT, was associated with a significantly decreased risk of glioma. The rs10764901 AA genotype carriers had an OR of 0.49 (95% CI, 0.24–0.98; P=0.045) compared with the rs10764901 GG genotype. When the rs10764901 AG and AA genotypes were pooled for analysis, a significantly decreased risk of glioma was also found (OR, 0.63; 95% CI, 0.43–0.93; P=0.021). Functional analyses showed that the rs10764901 A allele drove a lower luciferase expression and had higher transcription factor binding affinity than the G allele.
Conclusion: An enhancer variant of MGMT rs10764901 affects the regulatory activity of enhancer by altering the binding affinity of transcription factors and is associated with glioma susceptibility.
Keywords: glioma, MGMT, enhancer, genetic variation, susceptibility
Introduction
O6-methylguanine-DNA methyltransferase (MGMT) is an important DNA repair enzyme that plays a crucial role in protection against the carcinogenic effects of alkylating agents.1 MGMT transfers mutagenic and cytotoxic alkyl groups from DNA to its active site and then loses activity irreversibly.2 Therefore, a certain amount of MGMT is required to maintain its DNA repair activity. Previous studies have shown that aberrant expression of MGMT plays an important role in carcinogenesis.3–5 The increased frequency of oncogene mutations might be the pivotal carcinogenic mechanism of MGMT inactivation.6,7 On the other hand, the MGMT level was also reported to be associated with response to alkylating drugs in cancer.8,9 Promoter methylation, which is the most frequent mechanism of inactivation of MGMT in glioma,10 has been well established as a major prognostic factor and predictor of chemotherapy response for glioma.11,12 However, the expression level of MGMT is not exactly matched to the promoter methylation status,13,14 which indicates that other mechanisms also play a non-negligible role in MGMT expression regulation.
Some studies have revealed that several genetic variations of MGMT affect gene expression and then play an important role in carcinogenesis and progression of several cancers.15–17 For example, the T allele of a promoter genetic variation rs16906252 was found to reduce expression of MGMT and be associated with increased risk of MGMT-methylated colorectal cancer and improved survival of patients with glioma.15,17 In recent years, some novel significant genetic susceptibility factors of cancers have been identified by unbiased genome-wide association studies (GWAS). It is worthy to note that most of them are located in non-coding regions of genome.18 Furthermore, a number of non-coding genetic variations identified by GWAS occur in regions that act as enhancers and have functional effects on enhancer activity.18–20
In order to survey the role of enhancer variants of MGMT in glioma, we investigated their associations with glioma susceptibility and explored their functional relevance. Our data demonstrate that an intronic variant of MGMT rs10764901 affects the regulatory activity of an enhancer by altering the binding affinity of transcription factors and is associated with glioma susceptibility.
Patients and methods
Selection of candidate genetic variations
Several tracks of Ensembl Genome Browser (http://www.ensembl.org/index.html, Release 75) were used to select candidate genetic variations.21 First, we used the VISTA track to explore potential enhancers of MGMT.22 Not only the gene body of MGMT but also its 100 kb upstream and downstream flanking regions were analyzed. Then, the Genomic Evolutionary Rate Profiling (GERP) conservation scores based on the 37 eutherian mammals were used to measure the evolutionary conservation of sequences of the potential enhancers of MGMT.23 In order to screen the common genetic variations in our study population, the 1000 Genomes Project East Asian (1 KG EAS) common short variants (single-nucleotide polymorphisms [SNPs] and indels) track, which displays the variants with minor allelic frequency (MAF) >1% genotyped in East Asian individuals by the 1000 Genomes project, was used to analyze the variants located in the potential enhancers of MGMT.24 Because of the relatively small sample size, only the common variants with MAF >10% in Southern Han Chinese (CHS) population were selected for genotyping in this study.
Study subjects
This study recruited 150 incident glioma patients and 327 healthy controls. A portion of patients and controls was enrolled in our previous studies on glioma.25,26 In the present study, we extended the sample size of glioma patients from 138 to 150. All subjects were unrelated CHS living in Fujian, Zhejiang, and surrounding provinces. Patients with histopathologically confirmed glioma were recruited from January 2010 to October 2014 at the First Affiliated Hospital, Fujian Medical University (Fuzhou, n=122) and the First Affiliated Hospital, College of Medicine, Zhejiang University (Hangzhou, n=28). Healthy controls were cancer-free individuals recruited during the same period. At recruitment, written informed consent was obtained from each subject, and the detailed information on demographic and clinical characteristics were collected. This study was approved by the Institutional Review Board of the First Affiliated Hospital, Fujian Medical University.
Genotype analysis
Genomic DNA samples of 138 glioma patients and 327 healthy controls enrolled in our previous studies were isolated from the peripheral blood lymphocytes using a commercial Tiangen TIANamp Genomic DNA kit (Tiangen Biotech., Beijing, China).25,26 To increase the sample size of cases, 8.0% (12/150) of genomic DNA samples of cases were isolated from paraffin-embedded normal tissue adjacent to cancer specimens using KAPA Express Extract Kits (KAPA Biosystems, Wilmington, MA, USA). Genotypes of the candidate enhancer variants were determined by the Sequenom MassARRAY iPLEX platform (Sequenom Inc., San Diego, CA, USA). The information of primers is shown in Table S1. To control the data quality, genotyping was performed without knowledge of the case/control status of the subjects. And a 5% random sample was tested twice by different persons, and the reproducibility was 100%.
Functional analyses of genetic variation
In the present study, we performed functional studies only for rs10764901 which was identified to be associated with glioma susceptibility significantly.
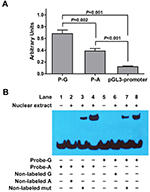
For luciferase reporter gene assays, two 1,366-bp DNA fragments corresponding to the potential enhancer (hs589) and containing rs10764901 G or A allele were amplified by polymerase chain reaction using same DNA sample template (primers are available upon request) and subcloned into the pGL3-promoter vector (Promega Corporation, Fitchburg, WI, USA). The resultant plasmid that contains rs10764901 G allele was designated as P-G, while the other one was designated as P-A. The two constructs were identical except for the different allele at rs10764901 polymorphic site. They were restriction mapped and sequenced to confirm their authenticity. U251 cell was used for luciferase assays. The constructed reporter plasmid or the blank pGL3-promoter plasmid was co-electrotransfected with pRL-TK (Promega Corporation) to U251 cell, using Celetrix Electroporator (Celetrix Biotech., Manassas, VA, USA). Three independent transfection experiments were done, and each was performed in triplicate. The luciferase activity was analyzed by a Dual-Luciferase Reporter Assay System (Promega Corporation).
For electrophoretic mobility shift assays (EMSA), synthetic double-stranded and 3’ biotin-labeled oligonucleotides corresponding to rs10764901 G (Probe-G) or A (Probe-A) sequences and U251 cell nuclear extract were incubated at 25°C for 20 minutes using the Light Shift Chemiluminescent EMSA Kit (Pierce, Rockford, IL, USA). For competition assays, non-labeled oligonucleotides at 150-fold molar excess were added to the reaction mixture before the addition of biotin-labeled probes. The reaction mixture was separated on 7% polyacrylamide gel electrophoresis, and the products were detected by Stabilized Streptavidin-Horseradish Peroxidase Conjugate (Pierce). Densitometric analyses of EMSA images were performed by ImageJ software (version 1.50i, NIH). The information of probes is shown in Table S2.
Statistical analysis
The differences in sex and age between cases and controls were examined by two-sided chi-squared test and Wilcoxon rank sum test, respectively. The associations of the candidate enhancer variants with glioma susceptibility were estimated by ORs and their 95% CIs computed by multivariate logistic regression model and adjusted for sex and age. Bonferroni correction was used to adjust the P-value for multiple testing. Meanwhile, the false-positive report probability (FPRP) was also calculated to assess the significance as described by Wacholder et al.27 Because of the relatively small sample size, we set the significance value of FPRP at 0.50 and the prior probability at 0.10 in this study. The statistical power to detect an OR of 1.50 (or its reciprocal, 0.67) was used to calculate FPRP value. We also performed the subgroup analyses by sex, age, and WHO grade. A t-test was used to examine the differences in luciferase activity between P-G, P-A, and the blank pGL3-promoter. The differences in densitometries of DNA-protein complexes formed with Probe-A and Probe-G were also analyzed by t-test. These statistical analyses were implemented in Statistic Analysis System software (version 9.4, SAS Institute, Cary, NC, USA) and STATA statistical software (version 14.0; StataCorp LP, College Station, TX, USA). P-value of <0.05 was used as the criterion of statistical significance, and all statistical tests were two sided.
Results
We first used Ensembl Genome Browser to choose candidate variants located in the potential enhancers of MGMT. As shown in Figure 1, four potential enhancers (hs656, hs696, hs331, and hs589) located in the intron of MGMT were identified by VISTA track. The potential enhancers hs656 and hs331 were excluded from further analyses because of the absence of common variants with MAF >10% in CHS population. The remaining two potential enhancers hs696 and hs589 were shown to have extreme evolutionary sequence conservation by the GERP conservation scores track. Two common variants rs577227 (MAF=0.39) and rs10764901 (MAF=0.41) were found in the potential enhancers hs696 and hs589, respectively. They were both selected as candidate enhancer variants for further genotyping.
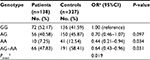
The detailed baseline characteristics of study subjects are presented in Table 1. There was no significant differences of age and sex distributions between glioma patients and controls, which suggested that the frequency matching of cases and controls was adequate. Of the 150 patients, 80 (53.3%) patients were classified into astrocytic tumors, 37 (24.7%) were classified into oligodendroglial tumors, 16 (10.7%) were classified into oligoastrocytic tumors, and 17 (11.3%) were other histological types. The detailed tumor WHO classification data were successfully collected from 136 (90.7%) patients. Among them, seven (4.7%) patients were WHO grade I, 45 (30.0%) were WHO grade II, 37 (24.7%) were WHO grade III, and 47 (31.3%) were WHO grade IV. There are no clear grade records for the remaining 14 (9.3%) patients. Maximal safe resection or subtotal resection were performed on all patients. Meanwhile, 101 (67.3%) and 112 (74.7%) patients also underwent radiotherapy- and alkylate-based chemotherapy, respectively.
  | Table 1 Selected characteristics of glioma patients and controls Notes: aWilcoxon rank sum test; bTwo-sided chi-squared test. |
The genotype distributions of rs10764901 and rs577227 in controls were in agreement with that expected under Hardy-Weinberg equilibrium. The multivariate logistic regression was used to investigate the associations between the candidate enhancer variants and glioma susceptibility with adjustment for sex and age (Table 2). As a result, a significant association between rs10764901 and glioma susceptibility was found. Subjects carrying rs10764901 AG or AA genotype had an adjusted OR of 0.67 (95% CI, 0.44–1.02; P=0.059) or 0.49 (95% CI, 0.24–0.98; P=0.045), respectively, compared with individuals carrying GG genotype. These results suggest that rs10764901 A allele may be a protective allele and acts in an allele dose-dependent manner (trend test; P=0.018). A more significant decreased risk of glioma was seen when groups of rs10764901 AG and AA genotype were pooled for analysis (adjusted OR, 0.63; 95% CI, 0.43–0.93; P=0.021). The association for rs10764901 was still significant after Bonferroni correction for two testings. Moreover, the estimated FPRP value for rs10764901 was 0.332 which is below the prespecified FPRP value of 0.50. It also suggests that our finding is noteworthy. We also examined whether genotype frequencies of rs10764901 differed between blood DNA and adjacent normal tissue DNA. No significant difference was found (Table S3). The significant association between rs10764901 and glioma susceptibility was also found when we repeated the association analyses without the 12 cases with adjacent normal tissue DNA (Table S4). These results verified that there was no significant heterogeneity in genotype frequencies for rs10764901 between different DNA sources. The other candidate enhancer variant rs577227 was not associated with glioma susceptibility significantly. Compared with subjects carrying rs577227 TT genotype, the adjusted ORs for TC or CC genotype were 1.33 (95% CI, 0.87–2.03; P=0.193) or 1.32 (95% CI, 0.69–2.53; P=0.410), respectively. When we combined rs577227 TC and CC genotype for analysis, the adjusted OR was 1.34 (95% CI, 0.89–2.01; P=0.156).
We then carried out subgroup analyses for rs10764901 based on sex, age, and WHO grade in a dominant model (Figure 2). The protective effect of rs10764901 A allele was observed across all subgroups, especially in males and patients with WHO grade III glioma. Significantly decreased risk of glioma was observed in males (adjusted OR, 0.53; 95% CI, 0.32–0.86; P=0.011). Similarly, it was found that rs10764901 was significantly associated with deceased risk of WHO grade III glioma (adjusted OR, 0.49; 95% CI, 0.25–0.99; P=0.046). No significant association was found in other subgroups although the trend is similar in all subgroups. It might be due to the small sample sizes within a stratum.
  | Figure 2 Subgroup analyses for rs10764901 based on sex, age, and WHO grade in a dominant model. Note: The central black dot represents the OR, and the horizontal line indicates the 95% CI. |
To investigate the regulatory activity of the potential enhancer hs589 and whether rs10764901 has a functional effect on the regulatory activity of hs589, we constructed two luciferase reporter gene plasmids (P-G and P-A) encompassing hs589 with rs10764901 G or A allele, respectively, and then transiently transfected into U251 cells. As shown in Figure 3A, about six- or threefold higher reporter gene expression was observed for the plasmid P-G or P-A, respectively, than the pGL3-promoter plasmid (P-G vs pGL3-promoter, P<0.001; P-A vs pGL3-promoter, P<0.001). These results suggest that hs589 might be a real functional enhancer element. Then, we compared the transcriptional activity of rs10764901 G and A alleles. The P-G construct drove about twofold higher luciferase expression than the P-A construct (P=0.002). This finding indicates that rs10764901 G>A change affects the enhancer activity of hs589.
To explore whether the differences in enhancer activity between rs10764901 G and A alleles are due to their ability to bind transcriptional activators or inhibitors, we conducted EMSA using U251 cell nuclear extract. As shown in Figure 3B, U251 cell nuclear extract was able to bind both Probe-G and Probe-A, and the binding patterns were similar. However, more DNA-protein complex was formed when Probe-A was incubated with U251 cell nuclear extract (Lane 4) compared to Probe-G (Lane 8) under the same experimental conditions, with the relative densitometries (mean ± SE) from three independent experiments being 1.08±0.06vs 0.73±0.07 (P=0.016). It indicates that the protein binding affinity of Probe-A was higher than that of Probe-G. In competition assays, 150-fold excess of non-labeled A or non-labeled G probe could completely eliminate the DNA-protein interaction (Lane 2 and Lane 6, respectively). Meanwhile, the DNA–protein interaction was partly eliminated by 150-fold excess of non-labeled mut probe (Lane 3 and Lane 7). These results indicate that the binding is sequence specific.
Discussion
In the present study, we demonstrated that an intronic variant of MGMT rs10764901 is significantly associated with glioma susceptibility by a retrospective case-control study involving 150 glioma patients and 327 controls. The rs10764901 A allele carriers were found to have a significantly decreased risk of glioma. Functional analyses revealed that the rs10764901 A allele had higher transcription factor binding affinity than the G allele and drove a lower luciferase expression.
MGMT is well known as a unique DNA repair gene that reverses DNA alkylation damage alone.1 It is widely considered to be a tumor suppressor gene because some studies have found that overexpression of MGMT is associated with decreased risk of several cancers.3,4 However, we found an interesting result that rs10764901 A allele, which drove a lower gene expression, significantly reduced the risk of glioma. It indicates that MGMT may function as an oncogene under certain circumstances. Consistent with our results, a study on colorectal cancer found that the T allele of MGMT promoter variant rs16906252 is associated with constitutively reduced gene expression and reduces the risk of MGMT-unmethylated colorectal cancer.17 These results indicate that MGMT overexpression may be a risk factor for MGMT-unmethylated cancer. Interestingly, a converse result that rs16906252 T allele is associated with an elevated risk of MGMT-methylated colorectal cancer was observed.17 These clues indicate that the role of MGMT in carcinogenesis is far more complex, and it might be completely opposite in cancer subclassifications with different methylation status of MGMT. Moreover, it was also reported that, in the context of different methylation status of MGMT, the prognostic effect of TERT gene is dichotomous,28 which supports the hypothesis that the mechanism of carcinogenesis is different for cancer subclassifications with different methylation status of MGMT. This postulation can partly explain the inconclusive results reported by previous published studies on MGMT genetic variations.29 The studies enrolling more patients with MGMT methylation might reach the opposite conclusion to those recruiting more MGMT-unmethylated patients.
Unfortunately, detailed information on methylation status of MGMT of glioma patients enrolled in this study is scant. Thus, we have no hard evidence to indicate that the MGMT-unmethylated glioma constituent ratio is higher than MGMT-methylated glioma in this study, which might partly explain why a deleterious role of MGMT overexpression in glioma carcinogenesis was inferred from our study. However, it is worthy to note that nearly two thirds of patients enrolled in this study are males. MGMT unmethylation has been found to be significantly higher in male patients with glioma.30,31 Therefore, it is plausible to deduce that MGMT-unmethylated glioma is in the majority in our study.
Still some other clues indicate that the physiological role of MGMT is far more than DNA repair. For instance, MGMT is expressed ubiquitously and its expression level far exceed its need to repair DNA damage.1 The exact role of phosphorylated MGMT is still an enigma.1 All of these and our findings indicate a broad function of MGMT needed further investigation.
A major limitation of this study is the relatively small sample size which might limit the statistical power. In addition, the lack of information on methylation status of MGMT results in the absence of subgroup analyses based on methylation status. Therefore, we could not explore directly whether rs10764901 has a converse role in developing glioma with different methylation status of MGMT.
Conclusion
We identified an enhancer variant of MGMT rs10764901 as a novel susceptibility locus for glioma for the first time. Our data revealed that rs10764901 A allele drove a lower gene expression and significantly reduced the risk of glioma. These results indicate that the underlying role of MGMT in glioma carcinogenesis needs further investigation. In addition, they warn us of the need for cancer risk stratification in study on genetic predisposition of cancer.
Acknowledgments
This study was supported by Joint Funds for the Innovation of Science and Technology, Fujian Province, China (grant no. 2016Y9016); Medical Elite Cultivation Program of Fujian Provincial Health and Family Planning Commission, China (grant no. 2016-ZQN-44); and National Natural Science Foundation, China (grant no. 81301772). The sponsors had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Author contributions
LH and XS designed this manuscript; LH, WX, LD, DY, and SZ conducted the study and collected glioma tissues and blood samples as well as corresponding clinical characteristics of cases and controls; LH and XS analyzed the data and wrote this manuscript. All authors contributed toward data analysis, drafting, and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4(4):296–307. | ||
Wibley JE, Pegg AE, Moody PC. Crystal structure of the human O(6)-alkylguanine-DNA alkyltransferase. Nucleic Acids Res. 2000;28(2):393–401. | ||
Liu L, Allay E, Dumenco LL, Gerson SL. Rapid repair of O6-methylguanine-DNA adducts protects transgenic mice from N-methylnitrosourea-induced thymic lymphomas. Cancer Res. 1994;54(17):4648–4652. | ||
Zhou ZQ, Manguino D, Kewitt K, et al. Spontaneous hepatocellular carcinoma is reduced in transgenic mice overexpressing human O6- methylguanine-DNA methyltransferase. Proc Natl Acad Sci U S A. 2001;98(22):12566–12571. | ||
Kawate H, Sakumi K, Tsuzuki T, et al. Separation of killing and tumorigenic effects of an alkylating agent in mice defective in two of the DNA repair genes. Proc Natl Acad Sci U S A. 1998;95(9):5116–5120. | ||
Nakamura M, Watanabe T, Yonekawa Y, Kleihues P, Ohgaki H. Promoter methylation of the DNA repair gene MGMT in astrocytomas is frequently associated with G:C --> A:T mutations of the TP53 tumor suppressor gene. Carcinogenesis. 2001;22(10):1715–1719. | ||
Esteller M, Toyota M, Sanchez-Cespedes M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60(9):2368–2371. | ||
Brent TP, Houghton PJ, Houghton JA. O6-Alkylguanine-DNA alkyltransferase activity correlates with the therapeutic response of human rhabdomyosarcoma xenografts to 1-(2-chloroethyl)-3-(trans-4-methylcyclohexyl)-1-nitrosourea. Proc Natl Acad Sci U S A. 1985;82(9):2985–2989. | ||
Schold SC, Brent TP, von Hofe E, et al. O6-alkylguanine-DNA alkyltransferase and sensitivity to procarbazine in human brain-tumor xenografts. J Neurosurg. 1989;70(4):573–577. | ||
Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59(4):793–797. | ||
Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. | ||
Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. | ||
Uno M, Oba-Shinjo SM, Camargo AA, et al. Correlation of MGMT promoter methylation status with gene and protein expression levels in glioblastoma. Clinics. 2011;66(10):1747–1755. | ||
Hsu CY, Lin SC, Ho HL, et al. Exclusion of histiocytes/endothelial cells and using endothelial cells as internal reference are crucial for interpretation of MGMT immunohistochemistry in glioblastoma. Am J Surg Pathol. 2013;37(2):264–271. | ||
McDonald KL, Rapkins RW, Olivier J, et al. The T genotype of the MGMT C>T (rs16906252) enhancer single-nucleotide polymorphism (SNP) is associated with promoter methylation and longer survival in glioblastoma patients. Eur J Cancer. 2013;49(2):360–368. | ||
Hsu CY, Ho HL, Lin SC, Ho TD, Ho DM. The MGMT promoter single-nucleotide polymorphism rs1625649 had prognostic impact on patients with MGMT methylated glioblastoma. PLoS One. 2017;12(10):e0186430. | ||
Kuroiwa-Trzmielina J, Wang F, Rapkins RW, et al. SNP rs16906252C>T Is an Expression and Methylation Quantitative Trait Locus Associated with an Increased Risk of Developing MGMT-Methylated Colorectal Cancer. Clin Cancer Res. 2016;22(24):6266–6277. | ||
Khurana E, Fu Y, Chakravarty D, Demichelis F, Rubin MA, Gerstein M. Role of non-coding sequence variants in cancer. Nat Rev Genet. 2016;17(2):93–108. | ||
Oldridge DA, Wood AC, Weichert-Leahey N, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528(7582):418–421. | ||
Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41(8):882–884. | ||
Flicek P, Amode MR, Barrell D, et al. Ensembl 2014. Nucleic Acids Res. 2014;42(Database issue):D749–D755. | ||
Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser – a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35(Database issue):D88–D92. | ||
Paten B, Herrero J, Beal K, Fitzgerald S, Birney E, Enredo BE. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18(11):1814–1828. | ||
1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. | ||
Huang L, Dai L, Xu W, Zhang S, Yan D, Shi X. Identification of expression quantitative trait loci of MTOR associated with the progression of glioma. Oncol Lett. 2018;15(1):665–671. | ||
Huang L, Xu W, Yan D, Dai L, Shi X. Identification of expression quantitative trait loci of RPTOR for susceptibility to glioma. Tumour Biol. 2016;37(2):2305–2311. | ||
Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434–442. | ||
Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics. 2017;14(2):284–297. | ||
Liu Y, Shete S, Hosking F, Robertson L, Houlston R, Bondy M. Genetic advances in glioma: susceptibility genes and networks. Curr Opin Genet Dev. 2010;20(3):239–244. | ||
Zawlik I, Vaccarella S, Kita D, Mittelbronn M, Franceschi S, Ohgaki H. Promoter methylation and polymorphisms of the MGMT gene in glioblastomas: a population-based study. Neuroepidemiology. 2009;32(1):21–29. | ||
Franceschi E, Tosoni A, Minichillo S, et al. The Prognostic Roles of Gender and O6-Methylguanine-DNA Methyltransferase Methylation Status in Glioblastoma Patients: The Female Power. World Neurosurg. 2018;112:e342–e347. |
Supplementary material
  | Table S1 Information of primers for Sequenom MassARRAY iPLEX assays |
  | Table S2 Information of probes for electrophoretic mobility shift assays |
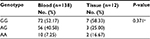  | Table S3 Genotype frequencies of rs10764901 genotyped using blood DNA or adjacent normal tissue DNA in glioma patients Notes: aTwo-sided χ2 test. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.