Back to Archived Journals » Hypoxia » Volume 7
An Erythrocytosis-Associated Mutation in the Zinc Finger of PHD2 Provides Insights into Its Binding of p23
Authors Song D, Guan W, Coon LM, Al-Kali A , Oliveira JL , Lee FS
Received 10 September 2019
Accepted for publication 16 October 2019
Published 13 December 2019 Volume 2019:7 Pages 81—86
DOI https://doi.org/10.2147/HP.S230502
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Dörthe Katschinski
Daisheng Song,1 Wei Guan,1–3 Lea M Coon,4 Aref Al-Kali,5 Jennifer L Oliveira,4 Frank S Lee1
1Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA; 2Department of Ultrasound, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, People’s Republic of China; 3Hubei Province Key Laboratory of Molecular Imaging, Wuhan 430022, People’s Republic of China; 4Hematopathology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA; 5Division of Hematology, Department of Internal Medicine, Mayo Clinic, Rochester, Minnesota, USA
Correspondence: Frank S Lee
Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, 605 Stellar Chance Bldg, 422 Curie Blvd, Philadelphia, PA 19104, USA
Tel +1215 898 4701
Fax +1215 573 2272
Email [email protected]
Background: Loss of function mutations in the EGLN1 gene are a cause of erythrocytosis. EGLN1 encodes for prolyl hydroxylase domain protein 2 (PHD2). PHD2 hydroxylates and downregulates hypoxia-inducible factor-2α (HIF-2α), a transcription factor that regulates erythropoiesis. While the large majority of erythrocytosis-associated EGLN1 mutations occur within its catalytic domain, rare mutations reside in its zinc finger. This zinc finger binds a Pro-Xaa-Leu-Glu motif in p23, an HSP90 cochaperone that facilitates hydroxylation of HIF-α, an HSP90 client. Essentially nothing is known about the specific interactions between the PHD2 zinc finger and p23.
Results: Here, we characterize an erythrocytosis-associated mutation in the zinc finger, K55N, that abolishes interaction with p23. We provide evidence that the affected residue, Lys-55, interacts with Asp-152 of p23. We also present results that indicate that PHD2 Arg-32 interacts with p23 Glu-160.
Conclusion: These studies not only reinforce the importance of the PHD2 zinc finger in the control of erythropoiesis, but also lead to a model in which a peptide motif in p23 binds in a specific orientation to a predicted groove in the zinc finger of PHD2.
Keywords: EGLN1, erythropoietin, hypoxia inducible factor, prolyl hydroxylase domain protein 2, polycythemia
Introduction
The HIF pathway critically regulates red cell mass.1–3 Under normoxic conditions, PHD2 constitutively site-specifically prolyl hydroxylates HIF-2α, providing a recognition motif for VHL, a component of an E3 ubiquitin ligase complex that targets HIF-2α for degradation.4–6 Under hypoxic conditions, this oxygen-dependent modification is arrested, leading to the stabilization of HIF-2α and activation of HIF-2α target genes, including that encoding for ERYTHROPOIETIN (EPO), the central hormone regulating red cell number. This allows the degree of tissue oxygenation to dictate the appropriate levels of red cell mass.
It is notable that mutations in the HIF pathway are a cause of erythrocytosis, a condition characterized by increased red cell mass.1,7 Specifically, loss of function mutations in EGLN1 (which encodes for PHD2), gain of function mutations in EPAS1 (which encodes for HIF-2α), or loss of function mutations in VHL are all associated with erythrocytosis. EPAS1 and VHL mutations are typically accompanied by increased EPO levels, while EGLN1-associated erythrocytosis often has normal EPO levels.8 In the case of EGLN1, the majority of mutations described reside within the catalytic domain. Where tested using biochemical and molecular biologic assays, these mutations produce a loss of function, i.e., a decreased capacity of PHD2 to hydroxylate HIF-2α.8
Subsequent to the discovery of catalytic domain mutations, we identified and characterized an erythrocytosis-associated loss of function mutation, C42R, in the zinc finger domain of the EGLN1 gene.9 This MYND-type zinc finger binds to a Pro-Xaa-Leu-Glu (PXLE) motif found in the HSP90 cochaperone p23.10 This promotes the recruitment of PHD2 to the HSP90 pathway and facilitates hydroxylation of HIF-2α, a known client of HSP90. The C42R mutation abolishes the interaction of PHD2 with p23 and likely disrupts the integrity of the zinc finger, since Cys-42 is predicted to be an essential zinc chelating residue.9 This mutation, therefore, does not yield information on the specific mechanism by which the PHD2 binds to p23. Indeed, there is essentially nothing known about the specific contacts made between p23 and PHD2.
Recently, we reported a different PHD2 zinc finger mutation, K55N, associated with erythrocytosis.11 Because it was not functionally studied, it was characterized as a variant of unknown significance. Here, we show that the mutation does, in fact, impair PHD2 function and report additional experiments which provide novel insights into PHD2 zinc finger binding to p23.
Materials and Methods
Plasmids
pcDNA3-HA-PHD2 (1–196), pcDNA5/FRT/TO-3xFlag-p23, pcDNA3-HA-p23, pEGFP-PHD2 (1–63) R32E, and pEGFP-PHD2 (1–63) R37E have been described.10,12
pcDNA3-HA-PHD2 (1–196) K55N was constructed as follows. A gene fragment containing the coding sequence of hemagglutinin (HA) tagged PHD2 (1–196) K55N was synthesized by IDT. The sequence of this gene fragment was: AGGGAGACCCAAGCTTGGTACCATGTACCCGTACGACGTGCCGGACTACGCAAGCGGGATCCGAATTCTTAAACTCGACATGGCTAATGATAGTGGTGGACCAGGTGGTCCTAGTCCAAGTGAGCGAGACCGGCAGTACTGTGAGCTCTGTGGTAAGATGGAGAACCTCTTAAGATGCAGTAGATGTAGGTCCTCGTTCTACTGCTGCAAGGAGCATCAGCGACAAGACTGGAAGAAGCACAATCTAGTATGTCAAGGTAGTGAAGGTGCTCTAGGCCATGGAGTTGGTCCACATCAACATTCTGGACCGGCCCCTCCGGCCGCAGTTCCACCACCTAGAGCTGGTGCACGTGAACCAAGAAAAGCAGCTGCTCGACGTGATAATGCTAGCGGAGATGCTGCTAAAGGAAAAGTAAAAGCTAAGCCTCCTGCAGATCCAGCTGCGGCCGCTTCACCTTGTAGAGCAGCCGCCGGTGGACAAGGATCTGCTGTAGCTGCAGAAGCTGAACCAGGTAAAGAAGAACCTCCTGCACGTTCGTCTCTATTTCAAGAAAAAGCTAATCTATATCCACCAAGTAACACTCCTGGAGATGCACTTAGTCCAGGAGGTGGACTACGTCCTAATGGACAAACAAAACCTCTTCCAGCTCTTAAACTAGCTCTCGAGTATCCCTATG. This DNA fragment was subsequently digested with Hind III/Xho I and subcloned into pcDNA3-HA generating the final vector pcDNA3-HA-PHD2 (1–196) K55N.
pcDNA3-HA-PHD2 (1–196) K55E was constructed in two steps. First, a gene fragment containing the coding sequence of hemagglutinin (HA) tagged wild type PHD2 (1–196) was synthesized by IDT. The sequence of this gene fragment was: AGGGAGACCCAAGCTTGGTACCATGTACCCGTACGACGTGCCGGACTACGCAAGCGGGATCCGAATTCTTAAACTCGACATGGCTAATGATAGTGGTGGACCAGGTGGTCCTAGTCCAAGTGAGCGAGACCGGCAGTACTGTGAGCTCTGTGGTAAGATGGAGAACCTCTTAAGATGCAGTAGATGTAGGTCCTCGTTCTACTGCTGCAAGGAGCATCAGCGACAAGACTGGAAGAAGCACAAGCTAGTATGTCAAGGTAGTGAAGGTGCTCTAGGCCACGGAGTTGGTCCACATCAACATTCTGGACCGGCCCCTCCGGCCGCAGTTCCACCACCTAGAGCTGGTGCACGTGAACCAAGAAAAGCAGCTGCTCGACGTGATAATGCTAGCGGAGATGCTGCTAAAGGAAAAGTAAAAGCTAAGCCTCCTGCAGATCCAGCTGCGGCCGCTTCACCTTGTAGAGCAGCCGCCGGTGGACAAGGATCTGCTGTAGCTGCAGAAGCTGAACCCGGGAAAGAAGAACCTCCTGCACGTTCGTCTCTATTTCAGAAAAAGCTAATCTATATCCACCAAGTAACACTCCTGGAGATGCACTTAGTCCAGGAGGTGGACTACGTCCTAATGGACAAACAAAACCTCTTCCAGCTCTTAAACTAGCTCTCGAGTATCCCTATG. This DNA fragment was subsequently digested with Hind III/Xho I and subcloned into pcDNA3-HA generating the vector pcDNA3-HA-PHD2 (1–196) WT. Secondly, the two following sequences were annealed: 5ʹ-GTCCTCGTTCTACTGCTGCAAGGAGCATCAGCGACAAGACTGGAAGAAGCACGAGCTCGTATGTCAAGGTAGTGAAGGTGCTCTAGGCCACGGAG-3ʹ and 5ʹ-GTGGCCTAGAGCACCTTCACTACCTTGACATACGAGCTCGTGCTTCTTCCAGTCTTGTCGCTGATGCTCCTTGCAGCAGTAGAACGAG-3ʹ. This oligonucleotide duplex was subcloned into the PpuM I/BstX I site of pcDNA3-HA-PHD2 (1–196) WT generating the final vector pcDNA3-HA-PHD2 (1–196) K55E.
pcDNA3-HA-PHD2 (1–196) K55D was constructed by subcloning into the PpuM I/BstX I site of pcDNA3-HA-PHD2 (1–196) WT a duplex consisting of the following two oligonucleotides: 5ʹ- GTCCTCGTTCTACTGCTGCAAGGAGCATCAGCGACAAGACTGGAAGAAGCACGATCTCGTATGTCAAGGTAGTGAAGGTGCTCTAGGCCACGGAG-3ʹ and 5ʹ- GTGGCCTAGAGCACCTTCACTACCTTGACATACGAGATCGTGCTTCTTCCAGTCTTGTCGCTGATGCTCCTTGCAGCAGTAGAACGAG-3ʹ.
pcDNA3-HA-PHD2 (1–196) R35E was constructed by subcloning an oligonucleotide duplex into the Afl II/PpuM I site of pcDNA3-HA-PHD2 (1–196) WT. The duplex consisted of the following two oligonucleotides: 5ʹ-TTAAGATGCAGTGAATGCAG-3ʹ and 5ʹ-GACCTGCATTCACTGCATC-3ʹ. pcDNA3-HA-PHD2 (1–196) R32E and pcDNA3-HA-PHD2 (1–196) R37E were constructed by overlapping PCR. In the first round of PCR, a 0.2 kb fragment was amplified using either pEGFP-PHD2 (1–63) R32E or pEGFP-PHD2 (1–63) R37E and the following primers: 5ʹ-CATGGTCCTGCTGGAGTTCGTG-3ʹ (EGFP C) and 5ʹ-GTGGCCGAGGGCGCCTTCACTACCTTGACATACA-3ʹ. A 0.4 kb fragment was also amplified using pcDNA3-HA-PHD2 (1–196) as a template and the following primers: 5ʹ-TGTATGTCAAGGTAGTGAAGGCGCCCTCGGCCAC-3ʹ and 5ʹ-TAGAAGGCACAGTCGAGG-3ʹ (BGH rev). The two PCR products were mixed and employed as a template in the second round of PCR using the EGFP C and BGH rev primers. The 0.6 kb product was digested with EcoR I/Xho I and subcloned into pcDNA3-HA.
pcDNA5/FRT/TO-3xFlag-p23 E160R was constructed by first PCR amplifying a 0.5 kb fragment using pcDNA3-HA-p23 as a template and the following two oligonucleotides: 5ʹ-GTACGGATCCAAATGCAGCCTGCTTCTGCAAAG-3ʹ and 5ʹ-GTACCTCGAGATATCTATCTCAGATCTGGCATTTTTTCATCATCAC-3ʹ. The product was cut with BamH I and Xho I and subcloned into pcDNA5/FRT/TO-3xFlag generating the final vector pcDNA5/FRT/TO-3xFlag-p23 E160R.
pcDNA5/FRT/TO-3xFlag-p23 D152K, pcDNA5/FRT/TO-3xFlag-p23 D153K, and pcDNA5/FRT/TO-3xFlag-p23 E154K were constructed by first amplifying a 0.5 kb fragment using pcDNA5/FRT/TO-3xFlag-p23 as a template, an oligonucleotide consisting of the sequence 5ʹ-GCGTGTACGGTGGGAGGTC-3ʹ, and an oligonucleotide consisting of one of the following sequences: 5ʹ-GTACCTCGAGCTTAAGTTACTCCAGATCTGGCATTTTTTCATCCTTACTGTCTTGTGAATCATCATC-3ʹ (D152K), 5ʹ- GTACCTCGAGTATCGATTACTCCAGATCTGGCATTTTTTCCTTATCACTGTCTTGTGAATCATCATC-3ʹ (D153K), or 5ʹ-GTACCTCGAGCCGCGGTTACTCCAGATCTGGCATTTTCTTATCATCACTGTCTTGTGAATCATCATC-3ʹ (E154K). The product was cut with BamH I and Xho I and subcloned into pcDNA5/FRT/TO-3xFlag.
The authenticity of all constructs was confirmed by Sanger sequencing.
Transfection and Immunoprecipitations
Cell culture, transfections, immunoprecipitations, and Western blotting were performed as previously described,10,12 except that in the case of coimmunoprecipitation experiments examining rescue of PHD2 (1–196) K55E or PHD2 (1–196) K55D binding to p23, the coimmunoprecipitation buffer consisted of 10 mM Tris, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.1% deoxycholate, 0.1% SDS (supplemented with Sigma mammalian cell protease inhibitor cocktail when used for cell lysis). For hypoxia experiments, cells were placed in a Heracell 150i Tri-gas incubator and exposed to 1% O2/5% CO2.
Results and Discussion
Genetic testing of patients with erythrocytosis for mutations in genes that include EGLN1, EPAS1, and VHL has revealed that some patients have pathogenic mutations in these genes, while others have variants of unknown significance.11 Included among the latter is a c.165G>C (p.K55N) mutation in the EGLN1 gene, found in a heterozygous state in two individuals.11
PHD2 Lys-55 is a conserved residue (Figure 1A, asterisk). The K55N mutation is not found in the Exome Sequencing Project, 1000 Genomes Project, or gnomAD. We examined the binding of PHD2 (1–196) K55N to p23 by a coimmunoprecipitation assay. Under conditions where wild type PHD2 (1–196) bound to p23, we found that the K55N mutation abolished it (Figure 1B, top panel, compare lanes 2 and 4). We cotransfected HEK293 cells with a Hypoxia Response Element (HRE) reporter gene along with expression constructs for wild type or K55N PHD2, exposed the cells to either normoxia or hypoxia, and then examined reporter gene activity. The HRE reporter gene contains three copies of the HRE from the human EPO gene enhancer.13 We observed that K55N PHD2 is weakened in its ability to downregulate HRE reporter gene activity (Figure 1C).
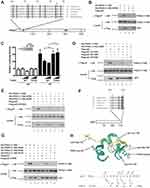 |
Figure 1 Functional characterization of K55N PHD2. (A) Diagram of PHD2, showing the location of zinc finger (ZF) and prolyl hydroxylase (PH) domains. Sequence of zinc finger across various metazoan species is shown at top. *= Lys-55 and += Arg-32 (human nomenclature). Shading = zinc chelating residues. (B, D, E, and G) HEK293FT cells were transfected with constructs for the indicated proteins. Cells were lysed, the Flag-tagged proteins were immunoprecipitated, and the immunoprecipitates examined for the absence or presence of HA-tagged PHD2 (1–196) by anti-HA Western blotting. Anti-HA and anti-Flag Western blots of lysates are also shown. Positions of molecular weight markers were as indicated. (C) HEK293FT cells in 96-well plates were transfected using Lipofectamine 2000 with 8 ng of (eHRE)3-Luc, 8 ng of RL-TK (which expresses Renilla luciferase under the control of the HSV thymidine kinase promoter), and either 0.8 or 2.5 ng of either pcDNA3-Flag-PHD2 or pcDNA3-Flag-PHD2 K55N. DNA doses were held constant by the addition of pcDNA3. Eight hr after transfection, cells were exposed to 1% O2 (HX) or maintained under normoxia (NX) for an additional 16 hr. All cells were lysed, and luciferase activities were measured and normalized to that of the Renilla luciferase internal transfection control. Shown are means ± SD, n = 3. **, p < 0.01 by student’s t-test. Anti-Flag Western blot of lysates of HEK293FT cells transfected with expression constructs for WT or K55N PHD2 is also shown. (F) Diagram of p23. ^= Asp-152 and # = Glu-160 (human nomenclature). Shading = P, L, and E of PXLE motif. (H) Top: model of p23 (152–160), which contains a PXLE motif, bound to the zinc finger of PHD2 (residues 20–59) generated as previously described12 and visualized using Protean 3 (DNASTAR). PHD2 is shown in green, p23 in yellow. Side chains of PHD2 Lys-55 and Arg-32, and p23 Asp-152, Pro-157, Leu-159, and Glu-160 are shown. Bottom: proposed contacts between residues in the C-terminal tail of p23 and residues in zinc finger of PHD2. N- and C-termini of the proteins are denoted by NH2 and COOH, respectively. |
Since lysine is a basic residue, this raises the possibility that it interacts with an acidic residue(s) in p23. Examination of a modeled three-dimensional structure of the PHD2 zinc finger bound to a p23 (152–160) peptide12 suggests that Lys-55 is a surface residue in the vicinity of the following acidic residues in p23: Asp-152, Asp-153, and Glu-154. We first mutated PHD2 Lys-55 to glutamic acid and find, as with the K55N mutation, that it abolished interaction with p23 (Figure 1D, top panel, compare lanes 2 and 4). We then tested whether the impaired interaction with p23 could be rescued by D152K, D153K, or E154K mutations in p23. We observed evidence of partial rescue with the D152K mutation (Figure 1D, top panel, lane 5) but not with the other mutations. The D152K mutation partially rescued a K55E, but not a K55D, mutation in PHD2 (Figure 1E, top panel, compare lanes 5 and 8), suggesting specificity for this rescue. Collectively, the data provide evidence that Lys-55 of PHD2 ordinarily contacts Asp-152 of p23.
These data also indicate that the p23 peptide binds in a specific orientation to the PHD2 zinc finger, and predicts that the C-terminal Glu of the PXLE motif (p23 Glu-160, Figure 1F) may be positioned in the vicinity of basic residues that include PHD2 Arg-32, Arg-35, and Arg-37. To test whether p23 Glu-160 forms such a critical ionic interaction, we first mutated p23 Glu-160 to Arg, and found that it abolished interaction with PHD2 (Figure 1G, top panel, compare lanes 2 and 3). We then examined the binding of p23 E160R to PHD2 R32E, R35E, and R37E, and found that only the first of these PHD2 mutations showed evidence of partial rescue of p23 E160R binding (top panel, compare lanes 6, 7, and 8). PHD2 R32E itself did not bind to wild type p23 (lane 5). These results, therefore, provide evidence that Glu-160 of p23 ordinarily forms a salt bridge with Arg-32 of PHD2.
Taken together, these findings strengthen the proposal that zinc finger mutations in PHD2 are a cause of erythrocytosis in humans. Importantly, these results now provide evidence for critical ionic interactions between PHD2 Lys-55 and p23 Asp-152, and PHD2 Arg-32 and p23 Glu-160 (Figure 1H, top). We found evidence for the rescue of a binding defect arising from a K55E PHD2 mutation by a p23 D152K mutation, and rescue of a binding defect arising from a p23 E160R mutation by an R32E PHD2 mutation. In both instances, the rescue was partial, perhaps due to unfavorable interactions between the mutated residues and neighboring amino acids on the same protein.
Mutagenesis and modeling studies indicate that additional critical interactions are likely contributed by the Pro and Leu of the PXLE motif, and Gln-47 and Trp-51 of PHD2.10,12 In the structure of the MYND-type zinc fingers of ETO and BS69 bound to PXL-containing peptides,14,15 the residues that correspond to Gln-47 and Trp-51 in PHD2 make contacts with the Pro of the PXL motif and the nonpolar residue immediately preceding this Pro, respectively. This suggests that Gln-47 and Trp-51 of PHD2 may, therefore, make similar contacts with p23 Pro-157 and Met-156, respectively (Figure 1H, bottom). In summary, these studies provide additional evidence for an important role for the PHD2 zinc finger in HIF regulation, and furthermore provide insights into how specificity is achieved in the interaction of PHD2 with its PXLE ligand.
Acknowledgments
This work was supported in part by NIH grants R01-DK104796 and R33-HL120751 to FSL. WG was supported by the Scientific Research Training Program for Young Talents (Union Hospital, Tongji Medical College, Huazhong University of Science and Technology).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lappin TR, Lee FS. Update on mutations in the HIF: EPO pathway and their role in erythrocytosis. Blood Rev. 2019;37:100590. doi:10.1016/j.blre.2019.100590
2. Haase VH. Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 2013;27(1):41–53. doi:10.1016/j.blre.2012.12.003
3. Wenger RH, Hoogewijs D. Regulated oxygen sensing by protein hydroxylation in renal erythropoietin-producing cells. Am J Physiol Renal Physiol. 2010;298(6):F1287–F1296. doi:10.1152/ajprenal.00736.2009
4. Kaelin WG
5. Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi:10.1016/j.molcel.2010.09.022
6. Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi:10.1016/j.cell.2012.01.021
7. Lee FS, Percy MJ. The HIF pathway and erythrocytosis. Annu Rev Pathol. 2011;6:165–192. doi:10.1146/annurev-pathol-011110-130321
8. Gardie B, Percy MJ, Hoogewijs D, et al. The role of PHD2 mutations in the pathogenesis of erythrocytosis. Hypoxia (Auckl). 2014;2:71–90. doi:10.2147/HP
9. Sinnema M, Song D, Guan W, et al. Loss-of-function zinc finger mutation in the EGLN1 gene associated with erythrocytosis. Blood. 2018;132(13):1455–1458. doi:10.1182/blood-2018-06-854711
10. Song D, Li LS, Heaton-Johnson KJ, Arsenault PR, Master SR, Lee FS. Prolyl Hydroxylase Domain Protein 2 (PHD2) binds a Pro-Xaa-Leu-Glu Motif, linking it to the heat shock protein 90 pathway. J Biol Chem. 2013;288(14):9662–9674.
11. Oliveira JL, Coon LM, Frederick LA, et al. Genotype-phenotype correlation of hereditary erythrocytosis mutations, a single center experience. Am J Hematol. 2018;93:1029–1041. doi:10.1002/ajh.v93.8
12. Arsenault PR, Song D, Chung YJ, Khurana TS, Lee FS. The zinc finger of Prolyl Hydroxylase Domain Protein 2 is essential for efficient hydroxylation of hypoxia-inducible factor alpha. Mol Cell Biol. 2016;36(18):2328–2343. doi:10.1128/MCB.00090-16
13. Yu F, White SB, Zhao Q, Lee FS. Dynamic, site-specific interaction of Hypoxia-inducible factor-1alpha with the von Hippel-Lindau tumor suppressor protein. Cancer Res. 2001;61(10):4136–4142.
14. Liu Y, Chen W, Gaudet J, et al. Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO’s activity. Cancer Cell. 2007;11(6):483–497. doi:10.1016/j.ccr.2007.04.010
15. Harter MR, Liu CD, Shen CL, et al. BS69/ZMYND11 C-terminal domains bind and inhibit EBNA2. PLoS Pathog. 2016;12(2):e1005414. doi:10.1371/journal.ppat.1005414
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
