Back to Journals » International Journal of Nanomedicine » Volume 10 » Issue 1
Amyloids in solid-state nuclear magnetic resonance: potential causes of the usually low resolution
Authors Espargaró A, Busquets MA , Estelrich J, Sabate R
Received 27 May 2015
Accepted for publication 18 July 2015
Published 9 November 2015 Volume 2015:10(1) Pages 6975—6983
DOI https://doi.org/10.2147/IJN.S89385
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Alba Espargaró, Maria Antònia Busquets, Joan Estelrich, Raimon Sabate
Department of Physical Chemistry, School of Pharmacy, Institute of Nanoscience and Nanotechnology (IN2UB), University of Barcelona, Barcelona, Spain
Abstract: Amyloids are non-crystalline and insoluble, which imply that the classical structural biology tools, ie, X-ray crystallography and solution nuclear magnetic resonance (NMR), are not suitable for their analysis. In the last years, solid-state NMR (ssNMR) has emerged as an alternative tool to decrypt the structural signatures of amyloid fibrils, providing major contributions to our understanding of molecular structures of amyloids such as β-amyloid peptide associated with Alzheimer’s disease or fungal prions, among others. Despite this, the wide majority of amyloid fibrils display low resolution by ssNMR. Usually, this low resolution has been attributed to a high disorder or polymorphism of the fibrils, suggesting the existence of diverse elementary β-sheet structures. Here, we propose that a single β-sheet structure could be responsible for the broadening of the line widths in the ssNMR spectra. Although the fibrils and fibers consist of a single elementary structure, the angle of twist of each individual fibril in the mature fiber depends on the number of individual fibrils as well as the fibril arrangement in the final mature fiber. Thus, a wide range of angles of twist could be observed in the same amyloid sample. These twist variations involve changes in amino acid alignments that could be enough to limit the ssNMR resolution.
Keywords: amyloid, fibril, misfolding, β-structure, ssNMR, NMR, β-sheet
Introduction
Amyloid aggregation of misfolded proteins that escape the cellular quality-control mechanisms is a common feature of a wide range of debilitating and increasingly prevalent diseases such as Alzheimer’s, Parkinson’s, Huntington’s, and Creutzfeldt–Jakob’s diseases.1 Amyloid fibrils display a core region formed from repetitive arrays of β-sheets oriented perpendicular to fibril axis forming the known cross-β structure.2 These fibrils are non-crystalline and insoluble, and, for this reason, they cannot be analyzed by X-ray crystallography and solution nuclear magnetic resonance (NMR), the classical structural biology tools.3 The knowledge of three-dimensional structure of amyloids is critical for understanding the mechanism of self-assembling and the design of potential inhibitors. In the same sense, the structural basis of prion infectivity remains largely elusive, mainly because of the difficulty in obtaining atomic resolution of such structures.4 In the last decade, solid-state NMR (ssNMR) has emerged as an alternative tool to decrypt the structural signatures of amyloid fibrils, providing major contributions to our understanding on the molecular structures of amyloids such as β-amyloid peptide associated with Alzheimer’s disease or fungal prions among others.5 Despite the numerous attempts to obtain the atomic molecular structure of the majority of amyloid fibrils such as Aβ(1–40)6 or Aβ(1–42),7 α-synuclein,8 transthyretin,9,10 amylin,11 PrP,12 Ure2p,13,14 Sup35p15 by ssNMR, the low resolution of such spectra has limited the acquisition of their atomic structure. Only the atomic structure of a few number of amyloid peptides, ie, HET-s prion-forming domain (HET-s PFD)4,16–19 and short peptides of Aβ,20–22 Sup35p,23 or β2-microglobulin24 have been well resolved. Thus, it could be considered low-resolved ssNMR spectra when the line broadening and the resonance splitting of ssNMR data limits the atomic structure acquisition and well-resolved ssNMR spectra when the atomic structure can be elucidated from ssNMR data. Since amyloid conformation seems to be an omnipresent process in both eukaryotic and prokaryotic organisms,25 a detailed analysis of decrypted structures could provide essential information on these self-assembling processes. In this sense, we use in the present work the decrypted structure of HET-s PFD to propose the potential causes of the observed low resolution of the most amyloid aggregates when analyzed by ssNMR.
Since prions are infectious proteins assembled as amyloid or amyloid-like structures that have a self-perpetuating capacity in vivo and, thus, turn into pathological infectious agents or protein-based genetic elements,26–28 structural models are essential to understand the prion and amyloid motifs. In contrast to the majority of amyloids, the fibrils of HET-s PFD at neutral pH display well-resolved ssNMR spectra facilitating their atomic structural resolution. Since HET-s protein participates in a fungal self/nonself recognition process called heterokaryon incompatibility,29 the surprising high structural order of HET-s PFD fibrils has been hypothesized to be the consequence of the fact that HET-s has an evolved biological function.5 The HET-s protein displays a globular α-helical domain appended to a natively unfolded PFD that act together in a structural model wherein amyloid fibrils built from a cross-β core are decorated with globular domains.30 The HET-s PFD structure based on ssNMR represents the first complete structure decrypted of an amyloid fibril. HET-s PFD fibrils assembled at neutral pH forms a left-handed β-solenoid, with each molecule forming two helical windings, and a highly ordered triangular cross-β compact hydrophobic core with three salt bridges and two asparagine ladders.4,16,19 This model is supported by electron diffraction where a prominent meridional reflection at 0.47 nm−1 (indicative of cross-β structure) is observed; moreover, scanning transmission electron microscopy measurements yielded 1.02±0.16 subunit per 0.94 nm, such values in agreement with the predicted value in the model of the fibrils formed at neutral pH.31
ssNMR spectra of HET-s PFD amyloids
A diversity of HET-s PFD amyloids formed in vitro has been reported.31,32 These fibrils, which share a width of 5 nm, are observed as tightly twisted fibrils at low pH and as bundled or disordered ones at neutral pH; moreover, whereas HET-s PFD fibrils formed at low pH are not infectious, all HET-s fibrils formed at neutral pH act as prions.32 The ssNMR spectra of uniformly 15N, 13C-labeled, fully hydrated HET-s PFD amyloid formed at neutral pH4,16,19 are qualitatively different from the spectra of Aβ(1–40),6 amylin,11 Ure2p(1–89) fibrils13 or HET-s PFD fibrils formed at low pH33 obtained under very similar conditions. The spectra of HET-s PFD fibrils formed at neutral pH exhibit 15N and 13C magic angle spinning (MAS) NMR line widths similar to those obtained for small proteins in microcrystalline states.34,35 No evidence for multiple sets of NMR chemical shifts has been reported for neutral pH HET-s PFD fibrils, suggesting the absence of polymorphism at the molecular level.5 In contrast, HET-s PFD fibrils at low pH display a spectral resolution with line widths between 128 and 202 Hz, which is somewhat worse than that shown by neutral pH fibrils wherein the values of 100 Hz are obtained. This lower resolution of HET-s PFD fibrils at low pH could be associated with a higher disorder degree (Figure 1).33
  | Figure 1 Proton-driven spin diffusion spectra of HET-s PFD fibrils formed at pH 3 (blue) and at pH 7 (red). |
Classical view for low ssNMR resolution of amyloid fibrils: polymorphisms at molecular level
Nowadays, amyloid fibril polymorphism is one of the main arguments to explain the low resolution obtained by ssNMR.5,36,37 Amyloid polymorphism, which is being reported for an increasing number of amyloid-prone proteins, probably reflects that fibrils have not been under evolutionary constraints to retain a single active conformation.38 Amyloid folding is a consequence of a delicate balance between a multitude of conformational states and of the inter-conversion between them in a complex network of equilibriums.39 In this context, the alternative conformations of amyloid proteins depend on their multi-step assembly pathways that could be altered by fibrillation conditions.40 In this way, it is widely accepted that the same amyloid sample could contain several amyloid structures with different β-sheet cores because of alternative amyloid pathways. The presence of different β-sheet cores could be the reason of the low-resolution ssNMR spectra. However, amyloid formation exhibits two classes of polymorphism: one referred to the metastable intermediates commonly observed during amyloid formation and the other, the range of conformational distinct mature fibrils often seen at the reaction endpoint.41 Hence, polymorphism at the molecular and/or the macroscopic level should be taken into account.
Challenging the classical view for the low ssNMR resolution of amyloids: the HET-s PFD case
The electronic interactions and the ionization of amino acid residues are the key factors in the formation of different types of amyloid fibrils with alternative β-sheet folds. Since the experimental isoelectric point of HET-s PFD is ~5,42 pHs below 4 and over 6 could switch between both alternative amyloid pathways, and non-prion and prion pathways, respectively. At neutral pH, acidic (aspartic and glutamic acids) and basic residues (arginine and lysine) are fully ionized entailing the formation of three salt bridges,19 that could be critical for neutral pH fibril assembling in β-solenoid with a triangular hydrophobic core. In contrast, at low pH, the lag of the salt bridges and the change in the ionic states of several amino acidic residues may arise from a perturbation in the subunit fold, although maintaining a rigid part almost exclusively in β-sheet conformation but – in contrast to the neutral pH fibrils – without flexible residues.33 These compiled evidences suggest that the pH acts as a trigger between two amyloid pathways in the HET-s model suggesting the formation of only two elementary β-sheet structures: non-prion and prion states at low and neutral pH, respectively.
It is relevant to point out that prion HET-s PFD fibrils are assembling at neutral pH in bundled or disordered macroscopic structures at high and low ionic strength, respectively (Figure 2). In contrast, at low pH, non-prion HET-s PFD fibrils are shown as twisted fibers as main macroscopic structure (Figure 2).31,32 This observation suggests an apparently high polymorphism degree at the macroscopic level for both elementary structures, ie, non-prion and prion HET-s PFD amyloids. It is important to note that whereas the seemingly disordered prion HET-s PFD undergoes aggregation, showing two macroscopic states, ie, bundled and disordered fibrils, sometimes mixed at intermediate conditions;32 such aggregates exhibit well-resolved 15N and 13C MAS NMR line widths. This confirms the existence of a single elementary amyloid structure without any polymorphism.19 Contrarily, the apparently more ordered low pH fibers display broadening line widths suggesting polymorphism. Since polymorphism at the molecular level would not be envisaged for the samples assembled at pH 3 and 7, the polymorphism at macroscopic level should be responsible for the low resolution of non-prion HET-s PFD (assembled at low pH). Nevertheless, up to now, the reason why the polymorphism at the macroscopic level of prion HET-s PFD fibrils does not affect the resolution of ssNMR spectra remains still unresolved.
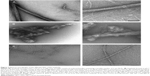  | Figure 2 Electron micrographs of the different HET-s PFD amyloids. |
Polymorphisms at macroscopic level: the effect of fibril maturation
ssNMR measurements require a total homogeneity of the sample to obtain a well-resolved spectrum. However, prion HET-s PFD fibrils at neutral pH, which usually display a large range of macroscopic structures, show surprisingly the well-resolved ssNMR spectra.19,31,32 This finding, indicative of single cross-β structure, suggests that bundled and disordered fibrils share the same cross-β motif. This fact suggests that both bundled and disordered HET-s PFD aggregates could be composed of single, independent, and identical fibrils, which, in turn, could be laterally associated with weak interactions forming stacks or disordered aggregates depending on the external conditions – low and high ionic strength. In this sense, when bundled and disordered HET-s PFD aggregates are brought to pH values close to 4–5 and sonication, neutral pH aggregates are gradually dissociated resulting in individual fibrils.31,32 Accordingly, detailed electronic microscopy analysis confirms the presence of 100% of individual fibrils into stacked aggregates of prion HET-s PFD fibrils at neutral pH.31 In contrast, non-prion HET-s PFD fibrils at pH 3.3 display three types of fibers – individual (92.7%), doublet (5.5%), and triplet (1.8%), and six types at pH 2.2 – doublet (0.8%), triplet (75.8%), quadruplet (0.1%), quintuplet (2.6%), sextuplet (18.4%), and septuplet (2.3%).31 Interestingly, these fibers cannot be easily dissociated by sonication.32 Thus, whereas only single fibrils should be considered at neutral pH, several non-dissociable macroscopic fibers are present at low pHs. In summary, at low pHs, single fibrils are strongly associated with forming fibers, whereas at neutral pH, the single fibrils are weakly associated probably with weak electrostatic forces remaining de facto as single fibrils. Interestingly, the polymorphism at macroscopic level has been reported for other amyloid-prone peptides as Aβ. Thus, the formation of several fibers of Aβ40 and Aβ42 displaying different pitch length and twist as well as several macroscopic associations has been shown by cryoEM.43–45 In the same way, several Aβ structures have recently resoluted by ssNMR.46,47 These compelling evidences suggest that macroscopic polymorphism could also be a potential cause of the usual low resolution of the Aβ ssNMR spectra.
There is a crucial difference between laterally associated single fibrils and single fibrils associated with forming fibers; whereas the laterally associated fibrils act as a solution of associated “cylinders”, the fibrils associated with forming fibers behave as a solution of associated “strands” of a rope (Figure 3). The main difference lies in the local fibril geometry when they approach each other. The effective “interaction area” between two “cylinders” is typically much larger than that for two “fibers”. This results in a much weaker attraction between “fibers” compared with that for “cylinders” association.48 Nevertheless, since fibers are vastly more stable structures than stacked cylinders, single fibrils associated with fibers are much more difficult to dissociate than those associated as stacked cylinders. In the HET-s model, HET-s PFD fibril stacks observed at neutral pH could be considered as an association of “cylinders” – considering that single fibrils act as a “cylinders”. A possible explanation is that finite fibrils are never stable if the primary twist strength is extremely – unrealistically – high; in this case, single fibrils are stable and infinite stacks are formed for higher inter-fibril sticking energies.48 At neutral pH, we observe protein precipitation – typical of this type of associations – forming different macrostructures that can be dissociated if the “inter-cylinder” stability is reduced (eg, pH, temperature, or sonication) obtaining the single fibrils. In contrast, low pH HET-s PFD fibrils form a clear and stable gel consistent with a veritable “fiber” system. Under these conditions, the inter-fiber association, entailing fiber stacking, is highly unlike because of the intrinsic morphology of the mature fibers, favoring the presence of non-precipitating fibers. In addition, the high fiber stability prevents the fiber dissociation and the existence of single fibrils.
  | Figure 3 Cylinders versus fibrils: three-dimensional views and some cross-sections. |
Amyloid mature fiber formation and amino acid alignments consequences
Amyloid fibrils are, for definition, repetitive and very ordered structures, and it is difficult to figure out, that these very regular structures can be heterogeneous and disordered as suggested by ssNMR. This essential paradigm could be explained for the model of self-assembling chiral rod-like units.49,50 A peptide in β-strand conformation could be considered as a chiral rod-like unit, with complementary donor and acceptor groups aligned on opposing sides, and having chemically different upper and lower surfaces. The rod-like “monomers” self-assemble via recognition between complementary donor and acceptor groups, to form long twisted tapes. The tape twist stems from the chirality of the monomers – naturally occurring amino acids are L-chirals – which gives rise to a left-handed twist around the long axis of the tape. The differences in the chemical structures of the two faces of the tape and in their affinity to the solvent lead to a cylindrical-like structure. One face of the tape is expected to be less soluble than the other. This chemical anisotropy results in inter-tape attraction and hence in double tape formation, obtaining single fibrils also named ribbons or proto-filaments or proto-fibrils when take part of a fiber. Both faces of the single fibrils are identical and are characterized by a saddle curvature. Hence, the single fibril does not bend, and its axis is straight at equilibrium (Figure 4).49,50 The sides of the single fibrils could be, in turn, mutually attracted resulting in stacks of cylinders or fibers in HET-s PFD model. Stacks are bundled or disordered aggregates, whereas fibers are twisted structures formed mainly by single fibrils triplets.
  | Figure 4 Single fibrils are antiparallel stacks monomers with varying geometries. |
The equilibrium structure of the individual “tapes” must exhibit a twist resulting from amino acid chirality,51 nevertheless, the fibrillogenesis from tape – non-stable specie – to single fibrils and fibers entails twist changes. In order to combine two tapes to form single fibrils, the twisted tapes must reduce their primary twist for inter-twist together. In the same manner, when the single fibrils are combined with fibers via weak face-to-face attraction, the stronger are the distortions of the single fibrils and more reduced is the twist-angle (Figure 5). The same is true if different fibers are joined together to form macro fibril structures. Additionally, the characteristics of each single fibril could determine the shape of the mature fiber in both the number of single fibrils and the position of the single fibrils in the rope. Interestingly, the amino acid composition could become a key factor in the fibril association determining the final twist-angle.52 Thus, importantly, there is a relationship between the number of single fibrils that forms a fiber and the twist reduction. In case of “small” peptides, the formation of a tape-to-tape union is essential for the single fibril formation. In contrast, “long” peptides, ie, Aβ(1–40) or HET-s PFD, have the tendency to form a roll structure in a same monomer obtaining single fibrils composed by a single subunit monomer per unit of length.3,31
  | Figure 5 Model of periodic multi-strand fibril produced by the aggregation of aperiodic single fibrils: fibrils of the nth order are twisted bundles containing n single fibrils. |
HET-s PFD single fibrils are composed by monomers in cross-β structure forming an axial stacking of β-solenoids with two coils per unit.31 HET-s PFD single fibrils at neutral pH are obtained as individual entities with the same angle of twist, entailing the same amino acid alignments. In contrast, HET-s PFD single fibrils at low pH are part of fibers mainly triplets and sextuplets. As previously stated, single fibrils in triplet or sextuplet structures display different twist-angle and consistently different amino acid alignments. Thus, at low pH, we should expect the existence of a single low pH HET-s PFD single fibril structure with different twists depending on both the number of single fibrils that compose every fiber and the macroscopic distribution of the single fibrils in the mature fiber (Figure 6). These amino acid alignment variations between consecutive and identical β-stand monomers could explain the low resolution of HET-s PFD fibers at low pH by ssNMR. In concordance of our assumption, recently it has been shown that spontaneous aggregation of the insulin-derived steric zipper peptide VEALYL resulting in different aggregation forms with common features. The comparison of ssNMR chemical shift data of twisted and untwisted structures from molecular dynamic (MD) simulations, suggest that different angles of twist could be the responsible for the experimentally observed line broadening and resonance splitting by ssNMR.53
  | Figure 6 Structural variability of HET-s PFD pH 2 fibers with multiple single fibrils. |
Conclusion
Amyloid aggregation can be considered as a universal and generic process. Since amyloid polymorphism has been reported for an increasing number of amyloid-prone proteins, it is likely that other amyloid-prone proteins display a similar behavior than that shown for HET-s PFD amyloids at low pH, showing different types of non-easily dissociable fibers composed for a discrete number of identical single fibrils with the same β-stand motif. The number of single fibrils as well as the relative position of those in the mature fiber could entail a wide range of angles of twist, involving changes in amino acid alignments between β-stand monomers that could be enough to limit the ssNMR resolution. The variation of the external conditions (eg, pH, temperature, ionic strength, agitation, and sonication) can alter the equilibrium among different amyloid structures favoring determined ones. The research of the ideal conditions to obtain a specific structure or the isolation of individual single fibrils will make possible better ssNMR determinations and open the possibility to decrypt new amyloid proteins in their amyloid fold.
Acknowledgments
RS is beneficiary of a contract under the Ramón y Cajal programme (RYC-2011-07987), and AE is beneficiary of a contract under the Juan de la Cierva programme (JCI-2012-12193), both financed by the Spanish Ministerio de Economía y Competitividad (MINECO). MAB and JE thank the financial support given by the MINECO to the project MAT2012-36270-C04-03. The authors thank the projects 2014SGR227 and 2014SGR938 of the Generalitat of Catalunya.
Disclosure
The authors report no conflicts of interest in this work.
References
Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. | ||
Carulla N, Caddy GL, Hall DR, et al. Molecular recycling within amyloid fibrils. Nature. 2005;436(7050):554–558. | ||
Thompson LK. Unraveling the secrets of Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2003;100(2):383–385. | ||
Ritter C, Maddelein ML, Siemer AB, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435(7043):844–848. | ||
Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q Rev Biophys. 2006;39(1):1–55. | ||
Petkova AT, Ishii Y, Balbach JJ, et al. A structural model for Alzheimer’s beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99(26):16742–16747. | ||
Luhrs T, Ritter C, Adrian M, et al. 3D structure of Alzheimer’s amyloid-beta(1–42) fibrils. Proc Natl Acad Sci U S A. 2005;102(48):17342–17347. | ||
Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci U S A. 2005;102(44):15871–15876. | ||
Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci U S A. 2004;101(3):711–716. | ||
Jaroniec CP, MacPhee CE, Astrof NS, Dobson CM, Griffin RG. Molecular conformation of a peptide fragment of transthyretin in an amyloid fibril. Proc Natl Acad Sci U S A. 2002;99(26):16748–16753. | ||
Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry. 2007;46(47):13505–13522. | ||
Lim KH, Nguyen TN, Damo SM, et al. Solid-state NMR structural studies of the fibril form of a mutant mouse prion peptide PrP89-143(P101L). Solid State Nucl Magn Reson. 2006;29(1–3):183–190. | ||
Baxa U, Wickner RB, Steven AC, et al. Characterization of beta-sheet structure in Ure2p(1)(-)(89) yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry. 2007;46(45):13149–13162. | ||
Chan JC, Oyler NA, Yau WM, Tycko R. Parallel beta-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry. 2005;44(31):10669–10680. | ||
Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci U S A. 2006;103(52):19754–19759. | ||
Siemer AB, Arnold AA, Ritter C, et al. Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc. 2006;128(40):13224–13228. | ||
Siemer AB, Ritter C, Ernst M, Riek R, Meier BH. High-resolution solid-state NMR spectroscopy of the prion protein HET-s in its amyloid conformation. Angew Chem Int Ed Engl. 2005;44(16):2441–2444. | ||
Siemer AB, Ritter C, Steinmetz MO, Ernst M, Riek R, Meier BH. 13C, 15N resonance assignment of parts of the HET-s prion protein in its amyloid form. J Biomol NMR. 2006;34(2):75–87. | ||
Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319(5869):1523–1526. | ||
Balbach JJ, Petkova AT, Oyler NA, et al. Supramolecular structure in full-length Alzheimer’s beta-amyloid fibrils: evidence for a parallel beta-sheet organization from solid-state nuclear magnetic resonance. Biophys J. 2002;83(2):1205–1216. | ||
Petkova AT, Buntkowsky G, Dyda F, Leapman RD, Yau WM, Tycko R. Solid state NMR reveals a pH-dependent antiparallel beta-sheet registry in fibrils formed by a beta-amyloid peptide. J Mol Biol. 2004;335(1):247–260. | ||
Bu Z, Shi Y, Callaway DJ, Tycko R. Molecular alignment within beta-sheets in Abeta(14–23) fibrils: solid-state NMR experiments and theoretical predictions. Biophys J. 2007;92(2):594–602. | ||
van der Wel PC, Lewandowski JR, Griffin RG. Solid-state NMR study of amyloid nanocrystals and fibrils formed by the peptide GNNQQNY from yeast prion protein Sup35p. J Am Chem Soc. 2007;129(16):5117–5130. | ||
Iwata K, Fujiwara T, Matsuki Y, et al. 3D structure of amyloid protofilaments of beta2-microglobulin fragment probed by solid-state NMR. Proc Natl Acad Sci U S A. 2006;103(48):18119–18124. | ||
de Groot NS, Sabate R, Ventura S. Amyloids in bacterial inclusion bodies. Trends Biochem Sci. 2009;34(8):408–416. | ||
Prusiner SB. Shattuck lecture – neurodegenerative diseases and prions. N Engl J Med. 2001;344(20):1516–1526. | ||
Caughey B, Baron GS. Prions and their partners in crime. Nature. 2006;443(7113):803–810. | ||
Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6(6):435–450. | ||
Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci U S A. 1997;94(18):9773–9778. | ||
Wasmer C, Schutz A, Loquet A, et al. The molecular organization of the fungal prion HET-s in its amyloid form. J Mol Biol. 2009;394(1):119–127. | ||
Sen A, Baxa U, Simon MN, et al. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked beta-solenoid model of HET-s prion fibrils. J Biol Chem. 2007;282(8):5545–5550. | ||
Sabate R, Baxa U, Benkemoun L, et al. Prion and non-prion amyloids of the HET-s prion forming domain. J Mol Biol. 2007;370(4):768–783. | ||
Wasmer C, Soragni A, Sabate R, Lange A, Riek R, Meier BH. Infectious and noninfectious amyloids of the HET-s(218–289) prion have different NMR spectra. Angew Chem Int Ed Engl. 2008;47(31):5839–5841. | ||
Castellani F, van Rossum BJ, Diehl A, Rehbein K, Oschkinat H. Determination of solid-state NMR structures of proteins by means of three-dimensional 15N-13C-13C dipolar correlation spectroscopy and chemical shift analysis. Biochemistry. 2003;42(39):11476–11483. | ||
Lange A, Becker S, Seidel K, Giller K, Pongs O, Baldus M. A concept for rapid protein-structure determination by solid-state NMR spectroscopy. Angew Chem Int Ed Engl. 2005;44(14):2089–2092. | ||
Eichner T, Radford SE. A diversity of assembly mechanisms of a generic amyloid fold. Mol Cell. 2011;43(1):8–18. | ||
Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu Rev Phys Chem. 2011;62:279–299. | ||
Pedersen JS, Dikov D, Flink JL, Hjuler HA, Christiansen G, Otzen DE. The changing face of glucagon fibrillation: structural polymorphism and conformational imprinting. J Mol Biol. 2006;355(3):501–523. | ||
Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. | ||
Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8(1):101–106. | ||
Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007;17(1):48–57. | ||
Sabate R, Lascu I, Saupe SJ. Characteristics of Thioflavin-T bound to HET-s amyloid fibrils assembled at pH 2. J Strcut Biol. 2008;162:387–396. | ||
Zhang R, Hu X, Khant H, et al. Interprotofilament interactions between Alzheimer’s Abeta1–42 peptides in amyloid fibrils revealed by cryoEM. Proc Natl Acad Sci U S A. 2009;106(12):4653–4658. | ||
Schmidt M, Sachse C, Richter W, Xu C, Fandrich M, Grigorieff N. Comparison of Alzheimer Abeta(1–40) and Abeta(1–42) amyloid fibrils reveals similar protofilament structures. Proc Natl Acad Sci U S A. 2009;106(47):19813–19818. | ||
Miller Y, Ma B, Tsai CJ, Nussinov R. Hollow core of Alzheimer’s Abeta42 amyloid observed by cryoEM is relevant at physiological pH. Proc Natl Acad Sci U S A. 2010;107(32):14128–14133. | ||
Xiao Y, Ma B, McElheny D, et al. Abeta(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22(6):499–505. | ||
Schutz AK, Vagt T, Huber M, et al. Atomic-resolution three-dimensional structure of amyloid beta fibrils bearing the Osaka mutation. Angew Chem Int Ed Engl. 2015;54(1):331–335. | ||
Nyrkova IA, Semenov AN, Aggeli A, Boden N. Fibril stability in solutions of twisted b-sheet peptides: a new kind of micellization in chiral systems. Eur Phys J B. 2000;17:481–497. | ||
Aggeli A, Nyrkova IA, Bell M, et al. Hierarchical self-assembly of chiral rod-like molecules as a model for peptide beta-sheet tapes, ribbons, fibrils, and fibers. Proc Natl Acad Sci U S A. 2001;98(21):11857–11862. | ||
Stroud JC, Liu C, Teng PK, Eisenberg D. Toxic fibrillar oligomers of amyloid-beta have cross-beta structure. Proc Natl Acad Sci U S A. 2012;109(20):7717–7722. | ||
Weatherford DW, Salemme FR. Conformations of twisted parallel beta-sheets and the origin of chirality in protein structures. Proc Natl Acad Sci U S A. 1979;76(1):19–23. | ||
Zappone B, De Santo MP, Labate C, Rizzuti B, Guzzi R. Catalytic activity of copper ions in the amyloid fibrillation of β-lactoglobulin. Soft Matter. 2013;9:2412–2419. | ||
Matthes D, Daebel V, Meyenberg K, et al. Spontaneous aggregation of the insulin-derived steric zipper peptide VEALYL results in different aggregation forms with common features. J Mol Biol. 2014;426(2):362–376. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
