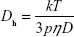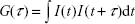Back to Journals » International Journal of Nanomedicine » Volume 11
Amphiphilic core shell nanoparticles containing dense polyethyleneimine shells for efficient delivery of microRNA to Kupffer cells
Authors Liu Z, Niu D, Zhang J, Zhang W, Yao Y, Li P, Gong J
Received 25 November 2015
Accepted for publication 17 March 2016
Published 15 June 2016 Volume 2016:11 Pages 2785—2797
DOI https://doi.org/10.2147/IJN.S101251
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Lei Yang
Zuojin Liu,1,* Dechao Niu,2,3,* Junyong Zhang,1 Wenfeng Zhang,1 Yuan Yao,2 Pei Li,2 Jianping Gong1
1Department of Hepatobiliary Surgery, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, 2Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hung Hom, Kowloon, Hong Kong, 3Lab of Low-Dimensional Materials Chemistry, School of Materials Science and Engineering, East China University of Science and Technology, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Abstract: Efficient and targeted delivery approach to transfer exogenous genes into macrophages is still a great challenge. Current gene delivery methods often result in low cellular uptake efficiency in vivo in some types of cells, especially for the Kupffer cells (KCs). In this article, we demonstrate that amphiphilic core–shell nanoparticles (NPs) consisting of well-defined hydrophobic poly(methyl methacrylate) (PMMA) cores and branched polyethyleneimine (PEI) shells (denoted as PEI@PMMA NPs) are efficient nanocarriers to deliver microRNA (miRNA)-loaded plasmid to the KCs. Average hydrodynamic diameter of PEI@PMMA NPs was 279 nm with a narrow size distribution. The NPs also possessed positive surface charges up to +30 mV in water, thus enabling effective condensation of negatively charged plasmid DNA. Gel electrophoresis assay showed that the resultant PEI@PMMA NPs were able to completely condense miRNA plasmid at a weight ratio of 25:1 (N/P ratio equal to 45:1). The Cell Counting Kit-8 assay and flow cytometry results showed that the PEI@PMMA/miRNA NPs displayed low cytotoxicity and cell apoptosis activity against the KCs. The maximum cell transfection efficiency reached 34.7% after 48 hours, which is much higher than that obtained by using the commercial Lipofectamine™ 2000 (1.7%). Bio-transmission electron microscope observation revealed that the PEI@PMMA NPs were mainly distributed in the cytoplasm of the KCs. Furthermore, when compared to the control groups, the protein expression of target nuclear factor κB P65 was considerably inhibited (P<0.05) both in vitro and in vivo. These results demonstrate that the PEI@PMMA NPs with a unique amphiphilic core–shell nanostructure are promising nanocarriers for delivering miRNA plasmid to KCs.
Keywords: branched polyethyleneimine, amphiphilic core–shell nanoparticles, Kupffer cells, gene delivery
Introduction
Kupffer cells (KCs), known as Browicz–Kupffer cells and stellate macrophages, are specialized macrophages located in the liver lining walls of the sinusoids that form part of the reticuloendothelial system. KCs account for 80%–90% of total fixed tissue macrophages in the body.1 Among all tissue-resident macrophages, KCs express a wide repertoire of receptors for the recognition of pathogens and danger signals such as Toll-like receptors, members of the inflammasome, and scavenger receptors.2,3 Using their special receptor repertoire, KCs are involved in liver physiological homeostasis and response to infection, toxins, transient ischemia, and various other stresses through the expression and secretion of multiple inflammatory mediators.4 Therefore, current investigations in KCs mostly focus on the signaling pathways that regulate inflammatory mediator production and subset development through all kinds of gene transfection methods.5
Although many techniques for expressing exogenous genes have been successfully developed in the past three decades, these methods are still not effective for the KCs as compared to other non-hematopoietic cells. This is due to the fact that macrophages are end-stage cells that generally do not divide. Therefore, those vectors that depend on integration into a replicative genome have only achieved limited success up to now. Furthermore, macrophages are quite responsive to “danger signal”. Thus, original viral vectors that have been used for gene transfer may induce potent antiviral responses in KCs, making these vectors not suitable for gene delivery.6 For example, adenovirus vectors of circulation are mainly eliminated by KCs and the innate immune responses mediated by adenoviruses-activated KCs can trigger robust proinflammatory cytokine release, such as interleukin (IL)-6 and tumor necrosis factor (TNF)-α.7,8
Alternatively, nonviral vectors, such as cationic liposomes, cationic polymer, synthetic peptides, and natural compounds, have attracted much attention due to their simple and controllable preparation as well as wide material selection.9,10 Among these nonviral carriers, branched polyethyleneimine (PEI) is considered as a promising polymer for gene delivery because of its highly positive charges and excellent complexing ability with the negatively charged nucleic acids through a strong electrostatic interaction. Moreover, after cellular internalization, the high amine density of branched PEI can facilitate the endosomal escape of the complexed DNA molecules via a well-known “proton sponge effect”, resulting in high transfection efficiency in a variety of cells.11 However, the use of PEI alone for gene delivery has several major drawbacks such as its cytotoxicity and broad size distribution of its polyplexes.12 Thus, developing an effective approach to reduce the toxicity of PEI without losing its high transfection efficiency is still highly desirable.
Several efforts have been devoted to overcome PEI cytotoxicity problem and enhance the transfection efficiency through modifying the PEI with biocompatible or even biodegradable components such as poly(ethylene glycol) (PEG), poly(L-lactide-co-glycolic acid) (PLGA), gelatin, and depsipeptide.13–18 For example, Lv et al reported that a complex micelle consisting of a biodegradable PLGA core and a mixed PEG/PEI shell can be prepared via the self-assembly of two block copolymers (PEG-b-PLGA and PLGA-b-PEI) in aqueous solution. This micelle can be used as a gene carrier for enhancing proliferation of endothelial cells.17 Recently, Li et al developed a new kind of gene carrier based on the amphiphilic block copolymers containing biodegradable hydrophobic depsipeptide copolymers and branched PEI.18 We have also developed a novel type of PEI-based amphiphilic core–shell nanoparticles (NPs) with diameters in the range of 100–300 nm and explored their potential application as nanocarriers for drug and gene deliveries.19–21 Compared with the abovementioned micellar-based biodegradable NPs, the PEI-based core–shell NPs possess several distinct advantages: 1) PEI@poly(methyl methacrylate) (PMMA) core–shell NPs with adjustable diameters ranging from 100 nm to 300 nm can be easily synthesized and scaled up; 2) the PEI shells allow rapid and efficient encapsulation of DNA molecules through the electrostatic complexation; 3) the immobilized PEI is three to four times less toxic than the native free PEI and is at least threefold more efficient as gene carriers in transfecting cells; 4) the amount of PEI needed to form complexed NPs with DNA molecules can be significantly reduced when using preformed uniform core–shell NPs containing PEI shells; and 5) the resulting DNA/NP complexes are highly uniform and homogeneous, providing good delivery system for pharmacokinetic studies. As part of our continuous effort in developing this type of PEI-based nanocarriers for drug and gene deliveries, this study aims at in vivo investigation of the PEI-based core–shell NPs in gene delivery, especially for transferring exogenous genes into KCs for future clinical applications. In this study, we have systematically evaluated the efficiency of PEI-based core–shell NPs as a nonviral gene vector to deliver nuclear factor κB microRNA (NF-κB-miRNA) plasmid to KCs in mice. The study included particle morphology, particle size, surface charge, complexing and releasing abilities of NF-κB-miRNA plasmid onto the NPs, and their cytotoxicity. Finally, the cellular uptake and miRNA delivery efficiency of the PEI-based core–shell NPs in KCs were also demonstrated both in vitro and in vivo.
Materials and methods
Synthesis of PEI@PMMA core–shell NPs
The PEI@PMMA NPs with core–shell structure were prepared according to our earlier established method.22,23 Briefly, the branched PEI (750 kDa, 50 wt% in water, 4.0 g) was first dissolved in water, and the resulting solution was then adjusted to pH 7.0 by adding 2 M HCl solution. The PEI solution was transferred into a water-jacketed flask equipped with a condenser, a thermocouple, a magnetic stirrer, and a nitrogen inlet, then stirred at 350 rpm, and purged with nitrogen for 30 minutes at 80°C. Purified methyl methacrylate monomer (8.0 g) was then added to the PEI solution. After mixing for 5 minutes, a dilute tert-butyl hydroperoxide solution (1 mL, 100 mM) was charged to the stirred mixture to initiate the polymerization, and the mixture was heated at 80°C for 2 hours under N2 atmosphere. After the reaction, stable particle dispersion was obtained, which was subsequently purified by centrifugation (15,000 rpm) for 15 minutes and washed with Milli-Q water. The purification cycle was repeated until conductivity of the supernatant was close to that of water used. Finally, the purified PEI@PMMA NP dispersion was stored at room temperature for subsequent use.
Characterization of PEI@PMMA NPs
The morphology was examined by field-emission scanning electron microscopy (SEM; JEOL-JSM 6335F) after coating the dried sample with a thin layer of gold under vacuum. Transmission electron microscopy (TEM) image was obtained using a JEOL-100-CXII TEM at an accelerating voltage of 100 kV. The sample was prepared by wetting a carbon-coated copper grid with a small drop of dilute particle dispersion in water (10 μL, 500 ppm). Upon drying, the NPs were stained with a small drop of 2 wt% phosphotungstic acid solution for an appropriate time and then dried at room temperature before TEM analysis. The hydrodynamic diameter and zeta-potential of the PEI@PMMA NPs were determined by using a Delsa™ Nano C (Beckman Coulter, Inc., Brea, CA, USA) analyzer with electrophoretic dynamic light scattering (DLS; a two-laser diode light source with a wavelength of 658 nm at 30 mW). Hydrodynamic diameter, Dh, was obtained from the Einstein–Stokes equation:
|
|
where k is the Boltzmann constant, η is the dispersant viscosity, T is the temperature (K), and D is the diffusion coefficient obtained from the decay rate of the intensity correlation function of the scattered light (ie, correlogram):
|
|
Surface charges of particles were measured based on their electrophoretic mobility. Samples for zeta-potential measurement were diluted to 100–200 ppm and measured at 25°C.
Construction of miRNA plasmid
The miRNA plasmid of NF-κB P65 was constructed by a vector pcDNA6.2-GW/EmGFP-miR (K4936-00; Thermo Fisher Scientific, Waltham, MA, USA). The forward chain of oligo DNA was 5′-TGCTGTTCAGTTGGTCCATTGAAAGGGTTTTGGCCACTGACTGACCCTTTCAAGACCAACTGAA-3′, while the reverse chain was 5′-CCTGTTCAGTTGGTCTTGAAAGGGTCAGTCAGTGGCCAAAACCCTTTCAATGGACCAACTGAAC-3′. The target sequence of miRNA was inserted into the sequence of 5′-GACTACACAAATCAGCGATTT-3′, which located at 3′-untranslated region of NF-κB P65. The plasmid was extracted by Omega Plasmid Extraction Kit (D6915-01; Omega).
Formation of PEI@PMMA/miRNA complexes
Various samples were prepared by mixing miRNA (0.5 mg/mL in phosphate-buffered saline [PBS], 0.01 M) and PEI@PMMA NPs (2 mg/mL in PBS, 0.01 M) at different weight ratios (30:1, 25:1, 20:1, 15:1, 10:1, 5:1, 2:1). The mixtures were all incubated at 37°C for 30 minutes on horizontal rotators. The resultant PEI@PMMA/miRNA complexes were loaded on 1% agarose gels. Gel electrophoresis was carried out at 100 V and 20 mA for 30 minutes. The migration patterns were obtained under a UV Gel Imager (Gel Doc™ XR; Bio-Rad Laboratories Inc., Hercules, CA, USA).
Animals
In our study, all C57/BL6 mice (male, 8 weeks old and 20–25 g) were purchased from the Animal Center of Chongqing Medical University. All mice were fed standard mouse food and housed in clear mice cages at 20°C. All in vivo procedures were approved by the Animal Care Committee of Chongqing Medical University. All animal-handling procedures were performed according to the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and followed the guidelines of the Animal Welfare Act.
Isolation and culture of mouse KCs
KCs were isolated from male C57/BL6 mice and then purified according to our previously published method.24 Briefly, mice were first anesthetized effectively, and portal vein was bluntly dissected and inserted with a sizeable catheter. Liver was excised and chopped after perfused in situ with PBS. The pieces of tissue were digested with 0.1% type IV collagenase at 37°C for 30 minutes in a water bath. The tissue homogenate was filtered using a 74 mm strainer following gradient density centrifugation for purification of KCs. Finally, the isolated cells were seeded in a dish and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; HyClone) and 1% penicillin/streptomycin (Pen/Strep) in a 5% CO2 atmosphere at 37°C. After culturing for 2–3 hours, the non-KCs were removed followed by washing with PBS. The remaining adherent cells, which proved to be mainly KCs, were continued to culture for 12–24 hours before different treatment.
Cytotoxicity of PEI@PMMA/miRNA complexes
The in vitro cytotoxicity of PEI@PMMA/miRNA complexes were studied by examining the survival rate of KCs using the Cell Counting Kit-8 assay (CCK-8) (C0038; Dojindo, Kumamoto, Japan). KCs in mice were first isolated and seeded in 96-well plates at a density of 1×105 cells/well and cultured for 24 hours. Following this, the PEI@PMMA/miRNA complexes with different weight ratios (30:1, 25:1, 20:1, 15:1, 10:1, 5:1, 2:1) were added into the culture medium containing 10% FBS. After incubation for 6 hours, 12 hours, 24 hours, and 48 hours, CCK-8 (10 μL, Dojindo) was added to each well and incubated at 37°C for 1 hour. The absorbance of each well was determined at 450 nm. Percent viability was defined as the relative absorbance of transfected versus blank control cells. Each final result was the average of six wells, and 100% viability was determined from untreated cells. The cell apoptosis was further confirmed by a flow cytometry (Gallio; Beckman Coulter). KCs were cocultured with gradient mass ratios (30:1, 25:1, 20:1, 15:1, 10:1, 5:1, 2:1) of PEI@PMMA/miRNA complexes for 48 hours, followed by digestion and resuspension, then determined by flow cytometry assay.
In vitro gene transfection of PEI@PMMA/miRNA complexes against KCs
PEI@PMMA NPs (40 μL, 2 mg/mL in PBS) and green fluorescent protein (GFP) containing miRNA plasmid (8 μL, 0.5 mg/mL in PBS) in PBS (52 μL) were mixed and incubated at 37°C for 0.5 hours. The mixture was added into a six-well plate containing KCs with 600 μL DMEM and 10% FBS and Pen/Strep as antibiotics. The fluorescence images were acquired under a Leica fluorescence microscope at different incubation times (12 hours, 24 hours, and 48 hours). The transfection rates were determined by a flow cytometry. For comparison, the commercial transfection agent Lipofectamine 2000 (Lipo2000) was used as a control.
Western blot analysis in vitro
KCs were planted in six-well plates and transfected with proper weight ratios of PEI@PMMA NPs or Lipo2000 and NF-κB-miRNA plasmid. Native PEI@PMMA NPs without plasmid and a blank group treated with the same culture medium were used as controls. The cells were harvested and extracted by 1× cell lysis (Beyotime, Haimen, Jiangsu, People’s Republic of China) buffer and prestained molecular weight markers (Beyotime) at 72 hours after transfection. Then, 30 μg/lane of cellular protein was electrophoresed through 10% sodium dodecyl sulfate–polyacrylamide gels and transferred to nitrocellulose membrane (Dako Denmark A/S, Glostrup, Denmark). Membranes were blocked in Tris-buffered saline (pH 7.5) with 0.5% Tween-20 and 5% defatted milk for 1 hour and then incubated with primary antibodies (NF-κB p65 monoclonal antibody, 1:500 dilution; Abcam, Cambridge, MA, USA). After incubation with horseradish peroxidase-conjugated secondary antibody (1:3,000 dilution; Boster, Wuhan, Hubei, People’s Republic of China), membranes were scored using the enhanced chemiluminescence (Boster) detection system (Amersham, Marlborough, MA, USA). A monoclonal antibody for β-actin (1:1,000 dilution; Beyotime, Haimen, Jiangsu, People’s Republic of China) was used as a control.
Bio-TEM analysis
KCs were first isolated from mice and seeded in six-well plates and cultured for 24 hours. The PEI@PMMA NPs (50 μL, 2 mg/mL in PBS) and GFP containing miRNA plasmid (8 μL, 0.5 mg/mL in PBS) in PBS (42 μL) were mixed and incubated at 37°C for 0.5 hours. The mixture was added into a six-well plate containing KCs with 600 μL DMEM and 10% FBS and Pen/Strep as antibiotics. After 6 hours of incubation, cultured cell samples were prepared on glass slides and fixed and solidified according to routine methods. After drying naturally, the samples were coated to these slides and observed under Bio-TEM. For the in vivo test, liver tissue samples were collected and fixed with 2.5% glutaraldehyde and 1% osmic acid. Tissues were then dehydrated with gradient concentrations of ethanol. Tissues were solidified with 100% acetone at 37°C under oven. They were then cut into slices at 50–60 nm thickness and stained with 3% uranyl acetate and lead citrate.
In vivo gene transfection of PEI@PMMA NPs
A total of 24 male C57/BL6 mice (8-week-old, 20–25 g) were involved in this study. The mice were randomly divided into three groups: the PEI@PMMA NPs/miRNA group, the Lipo2000/miRNA group, and the blank group. In the PEI@PMMA NPs/miRNA group, a mixture of 20 μg NF-κB-miRNA plasmid (0.5 μg/μL) and 0.5 mg PEI@PMMA NPs (20 μg/μL) was prepared for transfection through injecting into mouse liver via portal vein according to Piskounova et al.25 The Lipo2000/miRNA group also followed the same procedure, and the blank group was injected by normal saline as control. Then, half of the mice were sacrificed to obtain liver tissues for Bio-TEM analysis after 6 hours. The rest of the mice were finally sacrificed for evaluation of NF-κB protein levels with immunohistochemical (IHC) method and Western blot analysis. For IHC analysis, a sample of liver tissue measuring 1× 0.5 cm ×0.5 cm was collected and fixed by 10% formaldehyde for 24 hours. Then, the tissues were embedded with paraffin and cut into slices (5 μm). Then, the slides were deparaffinized by heating to 60°C for 15 minutes and then subjecting to two 5-minute changes in 100% xylene. The sections were then rehydrated by serial incubations in 100%, 90%, and 80% ethanol, followed by PBS and microwave for 15 minutes in 10 mM sodium citrate for antigen retrieval. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide and subsequently incubated in 5% goat serum to block nonspecific binding sites. The slides were then incubated at 4°C overnight with polyclonal antibody for NF-κB. The avidin–biotin technique was performed with a secondary biotinylated antibody and avidin–peroxidase complex kit (Boster). The slides were finally counterstained with hematoxylin. Target protein levels were evaluated through staining of tissues. All of these were analyzed with Image-Pro Plus 6.0 software (Media Cybernetics, Silver Spring, USA). For in vivo Western blot test, KCs were first extracted from mouse liver tissues as described earlier,24 and then the subsequent experimental procedure was consistent with that of cellular Western blot.
Enzyme-linked immunosorbent assay
Mouse plasma levels of IL-6 and TNF-α were evaluated with enzyme-linked immunosorbent assay (ELISA). ELISA assays were operated according to the manual of ELISA Assay Kit (4A Biotech, Beijing, People’s Republic of China). All solutions were also prepared according to the manual. A total of 100 μL samples or standard substance was added into the wells of the plate, and then 50 μL biotinylated working solution was added and mixed and incubated for 2 hours under room temperature. After removing all the liquid from the wells, the wells were washed four times with PBS. Then, 100 μL enzyme-bonded working solution was added and incubated for 30 minutes under room temperature and then washed four times. This was followed by addition of 100 μL coloring solution after which the reaction was terminated with a termination solution. Finally, OD450 was evaluated by UV spectrophotometer (Beckman Coulter).
Statistical analysis
The SPSS 18.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. All data are reported as mean ± SD. Data were analyzed by one-way analysis of variance (ANOVA) and, as indicated, subject to post hoc comparisons using two-tailed Student’s t-test. Differences were considered significant when P<0.05.
Results and discussion
Morphology and structural characterization of the PEI@PMMA NPs
Amphiphilic nanocarriers (denoted as PEI@PMMA NPs) consisting of well-defined PMMA cores and PEI shells were synthesized according to previous established methods.22,23 Then, the miRNA plasmid-loaded PEI@PMMA NPs (denoted as PEI@PMMA/miRNA NPs) were obtained by an electrostatic interaction between positively charged PEI shells of NPs and negatively charged miRNA plasmid. Morphology of PEI@PMMA NPs was observed with both SEM and TEM as shown in Figure 1A and B, respectively. Both SEM and TEM images show highly monodispersed spherical NPs with an average diameter of ~250 nm. To identify the nanostructure of the particles, we utilized the darker contrast provided by heavy atoms in TEM images as the probe. Since the amine-containing PEI molecules have much stronger binding affinity for heavy metal ions than the PMMA, staining the particle with a dilute phosphotungstic acid solution should provide the necessary contrast to display the placement of the hydrophilic PEI segments within the polymer particles. The inset of Figure 1B shows the TEM micrograph of the PEI@PMMA core–shell particles, which display well-defined regions: 1) the white center part is the region of PMMA polymer and 2) the darker shell is mainly composed of PEI, which is highly swelling in water. Results from these studies indicate that the PEI@PMMA NP has a well-defined core–shell nanostructure with thick PEI outer layer in water.
The colloidal stability and the surface charge of PEI@PMMA NPs were characterized by DLS and zeta-potential measurement. Results shown in Figure 1C indicated that the PEI@PMMA had an average diameter of 279 nm with a narrow size distribution (polydispersity index value of 0.027). This result is consistent with the large-scale SEM image (Figure 1A). Zeta-potentials of PEI@PMMA NPs at pH 7.4 were ~+35 mV (Figure 1D). The high-positive zeta-potential value at neutral pH was due to the presence of hairy PEI shell, which is essential to effectively condense the negatively charged miRNA plasmids and protect them against enzymatic degradation during in vivo circulation.
Binding ability of the PEI@PMMA core–shell NPs to condense NF-κB-miRNA plasmid
The binding ability of the PEI@PMMA core–shell NPs to condense NF-κB-miRNA plasmid and its optimal complexation amounts were determined by a gel electrophoresis assay. Figure 2 shows the results of PEI@PMMA/miRNA complexes at various mass ratios (30:1, 25:1, 20:1, 15:1, 10:1, 5:1, 2:1) of the NPs and miRNA plasmids. The naked miRNA was used as a control (lane P). The results indicated that complete complex formation between the PEI@PMMA NPs and miRNA plasmid was achieved at the NPs and miRNA plasmid mass ratio of 25:1 (N/P ratio equal to 45:1). To investigate the stability of the PEI@PMMA/miRNA complexes, we have prepared the complexes using N/P ratio of 45:1 and studied their complex stability in PBS solution up to 48 hours through monitoring their particle sizes with DLS. The results in Figure 3 showed that there is little change in average diameters up to 48 hours, indicating a good stability of the PEI@PMMA/miRNA complexes in physiological environment.
Cytotoxicity of PEI@PMMA/miRNA NPs on KCs
KCs were isolated from C57/BL6 mice and used as model cells for in vitro cytotoxicity study. Cytotoxicities of the PEI@PMMA/miRNA NPs with different mass ratios of NPs to miRNA were examined in KCs using CCK-8 assay (Figure 4). As shown in Figure 4A, the PEI@PMMA/miRNA complexes formed at different mass ratios all exhibit survival fraction higher than 90%, even at a high mass ratio of 30:1 and a long incubation time (up to 48 hours). These results suggested that the PEI@PMMA/miRNA-complexed NPs had low cytotoxicity to KCs. The negligible toxicity of the complexes may be attributed to the following two reasons: 1) anchoring PEI onto a supporting particle can effectively reduce the cytotoxicity of PEI since the immobilized PEI molecule is unable to form multiple attachments onto the negatively charged cell surface20,21 and 2) the positive charges of the PEI molecules can be shielded or neutralized when mixing with the negatively charged miRNA plasmid. To further confirm cell apoptotic rates induced by the PEI@PMMA/miRNA complexes, flow cytometry was employed using an Annexin V-fluorescein isothiocyanate/propidium iodide (FITC/PI) double-staining assay.26,27 Results in Figure 4B indicate that cell apoptosis rates of the KCs are 6.3%±0.5%, 9.3%±0.1%, 12.2%±0.5%, 12.4%±1.1%, 12.98%±0.3%, and 14.16%±0.39%, after treating with the PEI@PMMA/miRNA complexes at mass ratios of 5:1, 10:1, 15:1, 20:1, 25:1, and 30:1, respectively. Overall, the high survival percentage and the low cell apoptosis rate suggest that PEI@PMMA NPs are suitable nanocarriers for miRNA delivery into KCs in liver due to their low cytotoxicity.
Transfection efficiency of PEI@PMMA/miRNA system in vitro
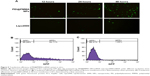
The transfection efficiency of PEI@PMMA/miRNA complexes in vitro was evaluated by transfecting the NF-κB-miRNA plasmid with GFP to KCs. To compare the effectiveness of PEI@PMMA NPs as gene carriers, a commercial transfection reagent, Lipo2000, was used as a control. Figure 5A illustrated fluorescent images of KCs observed after 12 hours, 24 hours, and 48 hours. The images show that increasing the incubation time from 12 hours to 48 hours results in significant increase in green fluorescence, indicating successful transfection of the PEI@PMMA/miRNA. On the contrary, very weak green fluorescence was observed in Lipo2000/miRNA group in all these incubation times. To further verify this effect, a flow cytometry analysis was employed. Figure 5B and C and showed that PEI@PMMA/miRNA exhibited transfection efficiency up to 34.7%. This is much higher than that of using commercial transfection agent Lipo2000 (1.7%). These results evidently suggest that the PEI@PMMA NPs are able to deliver the miRNA into KCs, while the Lipo2000 is not effective.
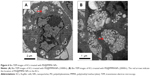
In order to confirm the entrance of PEI@PMMA NPs into KCs for delivering nucleic acid, the transfected KCs were observed with a Bio-TEM. The Bio-TEM images, as shown in Figure 6A and B, clearly display many white dots inside KCs, indicating the successful uptake of the PEI@PMMA NPs. Furthermore, the PEI@PMMA NPs mainly located in the cytoplasm, not into the nucleus of KCs. Thus, the PEI@PMMA NPs could effectively enter KCs for gene transfection.
Western blotting for target protein expressive level in vitro
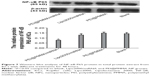
The inhibiting effect of cellular NF-κB p65 protein expression after treating with PEI@PMMA/miRNA complexes was studied by using Western blotting (WB). Figure 7 illustrates that PEI@PMMA/miRNA complexes could significantly inhibit NF-κB p65 protein expressions in the KCs, as compared with the use of only PEI@PMMA NPs group. These results indicated that PEI@PMMA NPs were able to deliver the miRNA plasmid into cells, and more importantly, the miRNA could self-replicate according to plasmid sequence and hampered the translation of target messenger RNA (mRNA), further limiting the formation of NF-κB p65 protein. In contrast, NF-κB p65 protein expression in the KCs treated with Lipo2000/miRNA was only slightly inhibited due to its low transfection ability as mentioned earlier. Based on the abovementioned results, the possible transfection mechanism of the PEI@PMMA/miRNA system on KCs is proposed as follows: through in vitro and in vivo circulation, the PEI@PMMA/miRNA complexes first entered the cells via an endocytosis and stayed in the lysosome, where it has lower pH value (5.0–5.5). The acidic environment in the lysosome facilitates endosomal escape and releases the miRNA in the cells through the well-known “proton sponge effect”.11 Subsequently, the intracellular miRNA was assembled into RNA, further identifying the target mRNA by complementary base pairing to hamper the translation of the target mRNA, resulting in silencing of NF-κB protein.
Transfection efficiency of PEI@PMMA/miRNA system in vivo
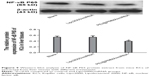
To investigate the transfection efficiency of PEI@PMMA/miRNA system in vivo, C57/BL6 mice were used as an animal model. In this study, PEI@PMMA/miRNA, Lipo2000/miRNA, and normal saline with the same volume as a blank group were injected into the mice via a portal vein.25 After 48-hour injection, the mice were sacrificed and the liver tissue was collected in order to target liver organ efficiently. Then, IHC analysis and WB were used to determine the NF-κB P65 protein levels in liver tissues of mice after different treatments. As shown in Figure 8, comparing to the Lipo2000/miRNA and blank groups, the PEI@PMMA/miRNA shows a significant reduction in brown color, indicating that the PEI@PMMA NPs could downregulate the expression of NF-κB P65 protein in mice liver tissues. WB results of KCs extracted from liver tissues (Figure 9) showed that the expression of NF-κB P65 protein was inhibited significantly with treatment of PEI@PMMA NPs/miRNA complexes when compared to other control groups. To further confirm the effectiveness of gene transfection to KCs in vivo, the tissue slices of liver selected from a mouse after treatment with PEI@PMMA/miRNA NPs were also characterized by Bio-TEM. Figure 10 illustrates that a large amount of PEI@PMMA NPs were located in the KCs of liver tissue, while a small portion of PEI@PMMA NPs were found in the vascular endothelial cell, revealing that PEI@PMMA/miRNA NPs could target the KCs in vivo. From the abovementioned results, it is concluded that PEI@PMMA NPs could induce targeted transfection to KCs in vivo; thus; they are promising nanocarriers for treatment of liver-related disease. The effective transfection is probably attributed to the special function of KCs, which could identify and engulf the miRNA-loaded PEI@PMMA NPs as foreign or harmful materials.28 Similar results have been reported by using other types of NPs.29,30 However, the detailed targeting mechanism of PEI@PMMA NPs still needs to be investigated in our further work.
Biological effect after the inhibition of NF-κB by the PEI@PMMA NPs/miRNA on KCs
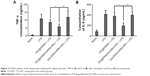
As reported earlier, the levels of proinflammatory cytokines such as TNF-α, IL-1β, IL-2, and IL-6 were significantly increased by activation of NF-κB signaling pathway.31 Thus, the silencing of NF-κB protein was an essential way to inhibit the release of proinflammatory cytokines.32 In the current work, we evaluated the biological effect of the PEI@PMMA NPs/miRNA on KCs. Two inflammatory mediators, TNF-α and IL-6, which were generated by KCs under the stimulation of lipopolysaccharide (LPS), were tested by ELISA. As shown in Figure 11A and B, the secretion of inflammation signal molecules TNF-α and IL-6 increased significantly after the stimulation of LPS, which is a typical inflammatory signaling pathway in KCs.33–35 In addition, compared with the PEI@PMMA NPs and Lipo2000/miRNA groups, both TNF-α and IL-6 levels in serum of mice were reduced significantly by inhibition of NF-κB P65 protein through miRNA-NF-κB transfection induced by PEI@PMMA NPs. On the other hand, Lipo2000 did not induce the decrease in these signal molecules due to its insufficient transfection to KCs. More importantly, no significant difference was observed between only LPS group and LPS + PEI@PMMA NPs group in the concentrations of signal molecules TNF-α and IL-6, indicating that PEI@PMMA NPs did not trigger the inflammation response of KCs. Therefore, our results indicate that PEI@PMMAs/miRNA NPs are capable of downregulating the secretion of inflammatory mediators from KCs and further confirm the effectiveness of PEI@PMMA NPs as gene delivery nanocarriers.
Conclusion
We successfully developed a new type of gene delivery system based on the core–shell-structured PEI@PMMA NPs for efficient delivery of miRNA plasmid to KCs. The nanocarrier of PEI@PMMA NPs displayed a well-defined core–shell structure, narrow size distribution, and high loading capacity with plasmid. Moreover, the transfection effect on the KCs of PEI@PMMA/miRNA evaluated both in vitro and in vivo showed that the PEI@PMMA/miRNA NPs exhibited low cytotoxicity and high transfection efficiency up to 34.7%. This transfection efficiency is much higher than that of the commercial transfection agent Lipo2000 (1.7%). Besides, the PEI@PMMA/miRNA NPs can transport into KCs efficiently and induce the expression and inhibition of NF-κB P65 in KCs in liver tissue, further resulting in the response of downstream signal molecules, such as TNF-α and IL-6. Therefore, the PEI@PMMA NP is a promising gene delivery carrier in the treatment of liver disease through delivering DNA or RNA into the KCs.
Acknowledgments
This research was supported by the Natural Science Foundation of China (No 81470899, 81177402), the National Scholarship Foundation of China (No 201208505116), the Outstanding Young Talent Fund of the Second Hospital of CQMU (2011), the Hong Kong Scholars Program (2014), China Postdoctoral Science Foundation (2014M560305), and Lo Ka Chung Centre for Natural Anti-Cancer Drug Development, The Hong Kong Polytechnic University.
Disclosure
The authors report no conflicts of interest in this work.
References
Jenne CN, Kubes P. Immune surveillance by the liver. Nat Immunol. 2013;14(10):996–1006. | ||
Stanculet N, Grigoras A, Avadanei R, Floarea-Strat A, Amalinei C, Caruntu ID. Relationship between Kuppfer cells, inflammation, and fibrosis in chronic hepatitis B and C. Rev Med Chir Soc Med Nat Iasi. 2013;117(4):880–889. | ||
Zeng WQ, Zhang JQ, Li Y, Yang K, Chen YP, Liu ZJ. A new method to isolate and culture rat Kupffer cells. PLoS One. 2013;8(8):e70832. | ||
Scholzel K, Schildberg FA, Welz M, et al. Transfer of MHC-class-I molecules among liver sinusoidal cells facilitates hepatic immune surveillance. J Hepatol. 2014;61(3):600–608. | ||
Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11(11):723–737. | ||
Zhang X, Edwards JP, Mosser DM. The expression of exogenous genes in macrophages: obstacles and opportunities. Methods Mol Biol. 2009;531:123–143. | ||
Seiradake E, Henaff D, Wodrich H, et al. The cell adhesion molecule “CAR” and sialic acid on human erythrocytes influence adenovirus in vivo biodistribution. PLoS Pathog. 2009;5(1):e1000277. | ||
Carlisle RC, Di Y, Cerny AM, et al. Human erythrocytes bind and inactivate type 5 adenovirus by presenting Coxsackie virus-adenovirus receptor and complement receptor 1. Blood. 2009;113(9):1909–1918. | ||
Oliveira C, Silveira I, Veiga F, Ribeiro AJ. Recent advances in characterization of nonviral vectors for delivery of nucleic acids: impact on their biological performance. Expert Opin Drug Deliv. 2015;12(1):27–39. | ||
Gu L, Nusblat LM, Tishbi N, et al. Cationic amphiphilic macromolecule (CAM)-lipid complexes for efficient siRNA gene silencing. J Control Release. 2014;184:28–35. | ||
Nel AE, Madler L, Velegol D, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8(7):543–557. | ||
Wu Y, Wang W, Chen Y, et al. The investigation of polymer-siRNA nanoparticle for gene therapy of gastric cancer in vitro. Int J Nanomedicine. 2010;5:129–136. | ||
Petersen H, Fechner PM, Fischer D, Kissel T. Synthesis, characterization, and biocompatibility of polyethylenimine-graft-poly (ethylene glycol) block copolymers. Macromolecules. 2002;35(18):6867–6874. | ||
Petersen H, Fechner PM, Martin AL, et al. Polyethylenimine-graft-poly (ethylene glycol) copolymers: influence of copolymer block structure on DNA complexation and biological activities as gene delivery system. Bioconjug Chem. 2002;13(4):845–854. | ||
Lee MS, Kim MG, Jang YL, et al. Polyethylenimine-g-poly (lactic-co-glycolic acid) as non-toxic micelle-type carrier for gene delivery. Macromol Res. 2011;19(7):688–693. | ||
Mimi H, Ho KM, Siu YS, Wu A, Li P. Polyethyleneimine-based core-shell nanogels: a promising siRNA carrier for argininosuccinate synthetase mRNA knockdown in HeLa cells. J Control Release. 2012;158(1):123–130. | ||
Lv J, Hao X, Yang J, Feng Y, Behl M, Lendlein A. Self-assembly of polyethylenimine-modified biodegradable complex micelles as gene transfer vector for proliferation of endothelial cells. Macromol Chem Phys. 2014;215:2463–2472. | ||
Li Q, Shi C, Zhang W, Behl M, Lendlein A, Feng Y. Nanoparticles complexed with gene vectors to promote proliferation of human vascular endothelial cells. Adv Healthc Mater. 2015;4(8):1225–1235. | ||
Ho KM, Li WY, Wong CH, Li P. Amphiphilic polymeric particles with core-shell nanostructures: emulsion-based syntheses and potential applications. Colloid Polym Sci. 2010;288(16–17):1503–1523. | ||
Zhu J, Tang A, Law LP, et al. Amphiphilic core-shell nanoparticles with poly (ethylenimine) shells as potential gene delivery carriers. Bioconjug Chem. 2005;16(1):139–146. | ||
Siu YS, Li L, Leung MF, Lee KL, Li P. Polyethylenimine-based amphiphilic core-shell nanoparticles: study of gene delivery and intracellular trafficking. Biointerphases. 2012;7(1–4):16. | ||
Li P, Zhu J, Sunintaboon P, Harris FW. New route to amphiphilic core-shell polymer nanospheres: graft copolymerization of methyl methacrylate from water-soluble polymer chains containing amino groups. Langmuir. 2002;18(22):8641–8646. | ||
Li W, Li P. Synthesis of well-defined amphiphilic core–shell particles containing amine-rich shells. Macromol Rapid Commun. 2007;28(23):2267–2271. | ||
Li PZ, Li JZ, Li M, Gong JP, He K. An efficient method to isolate and culture mouse Kupffer cells. Immunol Lett. 2014;158(1–2):52–56. | ||
Piskounova E, Polytarchou C, Thornton JE, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147(5):1066–1079. | ||
Henery S, George T, Hall B, Basiji D, Ortyn W, Morrissey P. Quantitative image based apoptotic index measurement using multispectral imaging flow cytometry: a comparison with standard photometric methods. Apoptosis. 2008;13(8):1054–1063. | ||
Brumatti G, Sheridan C, Martin SJ. Expression and purification of recombinant annexin V for the detection of membrane alterations on apoptotic cells. Methods. 2008;44(3):235–240. | ||
Strauss O, Dunbar PR, Bartlett A, Phillips A. The immunophenotype of antigen presenting cells of the mononuclear phagocyte system in normal human liver-a systematic review. J Hepatol. 2015;62(2):458–468. | ||
Poelstra K, Prakash J, Beljaars L. Drug targeting to the diseased liver. J Controll Release. 2012;161(2):188–197. | ||
Higuchi Y, Nishikawa M, Kawakami S, Yamashita F, Hashida M. Uptake characteristics of mannosylated and fucosylated bovine serum albumin in primary cultured rat sinusoidal endothelial cells and Kupffer cells. Int J Pharm. 2004;287(1–2):147–154. | ||
Fukada H, Yamashina S, Izumi K, et al. Suppression of autophagy sensitizes Kupffer cells to endotoxin. Hepatol Res. 2012;42(11):1112–1118. | ||
Blériot C, Dupuis T, Jouvion G, Eberl G, Disson O, Lecuit M. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity. 2015;42(1):145–158. | ||
Rameshrad M, Maleki-Dizaji N, Vaez H, Soraya H, Nakhlband A, Garjani A. Lipopolysaccharide induced activation of toll like receptor 4 in isolated rat heart suggests a local immune response in myocardium. Iran J Immunol. 2015;12(2):104–116. | ||
Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249(4975):1431–1433. | ||
Plociennikowska A, Hromada-Judycka A, Borzecka K, Kwiatkowska K. Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cell Mol Life Sci. 2015;72(3):557–581. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.