Back to Journals » Nature and Science of Sleep » Volume 14
Alterations of the Gut Microbiota in Response to Total Sleep Deprivation and Recovery Sleep in Rats
Authors Wang Z, Yuan K, Ji YB, Li SX , Shi L, Wang Z, Zhou XY, Bao YP, Xie W, Han Y, Shi J, Lu L, Yan W, Chen WH
Received 19 August 2021
Accepted for publication 31 December 2021
Published 25 January 2022 Volume 2022:14 Pages 121—133
DOI https://doi.org/10.2147/NSS.S334985
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Ahmed BaHammam
Zhong Wang,1,* Kai Yuan,1,* Yan-Bin Ji,2,* Su-Xia Li,3 Le Shi,1 Zhe Wang,1 Xin-Yu Zhou,4 Yan-Ping Bao,3 Wen Xie,5 Ying Han,3 Jie Shi,3 Lin Lu,1,3,6 Wei Yan,1 Wen-Hao Chen1
1Peking University Sixth Hospital, Peking University Institute of Mental Health, NHC Key Laboratory of Mental Health (Peking University), National Clinical Research Center for Mental Disorders (Peking University Sixth Hospital), Peking University, Beijing, 100191, People’s Republic of China; 2Department of Neurology, Qilu Hospital of Shandong University, Shandong University, Jinan, 250012, People’s Republic of China; 3National Institute on Drug Dependence and Beijing Key Laboratory of Drug Dependence, Peking University, Beijing, 100191, People’s Republic of China; 4Department of Psychiatry, The First Affiliated Hospital of Chongqing Medical University, Chongqing, 400016, People’s Republic of China; 5Mental Health Center of Anhui Province, Hefei, 230032, People’s Republic of China; 6Peking-Tsinghua Center for Life Sciences and PKU-IDG/McGovern Institute for Brain Research, Peking University, Beijing, 100871, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wen-Hao Chen; Wei Yan Peking University Sixth Hospital/Peking University Institute of Mental Health, 51 Huayuanbei Road, Haidian District, Beijing, 100191, People’s Republic of China
Tel +86-10-82801342
; Tel +86-10-62723704
Email [email protected]; [email protected]
Introduction: Accumulating evidence suggests that both sleep loss and gut dysbiosis can lead to metabolic disorders. However, less is known about the impact of total sleep deprivation (SD) and sleep recovery on the composition, function, and metabolic dynamics of the gut microbiota.
Methods: Specific-pathogen free Sprague–Dawley rats were subjected to 48 h of SD with gentle handling and then allowed to recover for 1 week. Taxonomic profiles of fecal microbiota were obtained at baseline, 24 h of SD, 48 h of SD, and 1 week of recovery. We used 16S rRNA gene sequencing to analyze the gut microbial composition and function and further characterize microbiota-derived metabolites in rats.
Results: The microbiota composition analysis revealed that gut microbial composition and metabolites did not change in the rats after 24 h of SD but were significantly altered after 48 h of SD. These changes were reversible after 1 week of sleep recovery. A functional analysis was performed based on Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations, indicating that 19 KEGG pathways were significantly altered in the gut microbiota in SD rats. These functional changes occurred within 24 h of SD, were more apparent after 48 h of SD, and did not fully recover after 1 week of sleep recovery.
Conclusion: These results indicate that acute total SD leads to significant compositional and functional changes in the gut microbiota, and these changes are reversible.
Keywords: gut microbiota, sleep deprivation, short-chain fatty acid, function prediction
Introduction
Sleep plays a critical role in physical and mental health maintenance and well-being throughout an individual’s lifetime. Sleep loss is typically associated with impairments in performance, negative health consequences, and lower survival.1,2 Sleep deprivation (SD) is a widespread phenomenon that is associated with adverse metabolic consequences,3–5 such as obesity, insulin resistance, diabetes, and cardiovascular disease, that ultimately promote the emergence of metabolic dysfunction for reasons that are still unclear.6
The gut microbiome is a vital component of the intestinal environment. It has been identified as a critical environmental factor that contributes to the development of obesity,7 insulin resistance,8 and diabetes.9 The gut microbiota-brain axis has become an increasingly popular research focus.10 Alterations of the intestinal microbiota have been linked to the pathophysiology of many psychiatric disorders, such as autism spectrum disorder,11 schizophrenia,12 and depression,13–15 suggesting that the gut microbiota affects multiple aspects of brain development and neuroendocrinological function. Research also demonstrates that microbiota-derived metabolites, such as short-chain fatty acids (SCFAs),16 are primarily produced in the large intestine through anaerobic bacterial fermentation and can maintain both intestinal immune function and gut barrier function.17 Accumulating evidence indicated that SCFAs played an important role in maintaining health and preventing disease.18 Recent studies showed that butyrate treatment led to rapid and robust increases in non-rapid-eye-movement (NREM) sleep in rats,19 highlighting the associations between sleep and gut microbial metabolites.
The gut microbiota fluctuates in response to circadian rhythm and routine schedules.20,21 A widely held presumption is that there is a relationship between sleep and the gut microbiome. Both sleep loss and dysbiosis of the gut microbiome are associated with metabolic diseases, such as obesity and diabetes.7,9,22 Several studies reported preliminary evidence that the gut microbiota was involved in sleep disorders in both murine models and human patients. Insomnia was shown to lead to significant structural and functional changes in the gut microbiota.23,24 A previous study found that the composition of the gut microbiota was subtly affected, with a higher Firmicutes/Bacteroidetes ratio in human subjects with sleep restriction.25 In contrast, recent work suggested that a single brief period of SD (5 h) did not alter the overall microbial composition.26 These preliminary studies highlighted the potential relationship between SD and changes in the gut microbiota. To date, no studies have comprehensively examined the impact of total SD and sleep recovery on the composition, function, and metabolic dynamics of the gut microbiota. The present study investigated the effects of acute total SD and 1 week of sleep recovery on microbiota populations. We used 16S ribosomal RNA (rRNA) gene sequencing to evaluate the compositional and functional changes in the gut microbiota in response to SD and sleep recovery.
Materials and Methods
Rat Sleep Deprivation and Sample Collection
Adult male specific-pathogen free Sprague-Dawley rats (7–8 weeks old) were housed individually under a 12 h/12 h light/dark cycle (lights on from 8:00 AM to 8:00 PM) and a controlled temperature of 21–22°C and humidity of 55% ± 5%. Standard laboratory irradiated chow food (Cobalt 60) and sterile drinking water were provided ad libitum. The rats (n = 8) were subjected to 48 h of SD and then allowed to recover for 1 week. The rats were in their home cages during SD. To reduce the rats’ stress, they were kept awake by gently tapping and shaking the cage by an experimenter.27,28 The control group (n = 8) had a regular sleep-wake cycle in the same environment (Figure 1A). The weight of the rats was recorded over the deprivation period. Each day, the same amount of food was provided to the SD and non-SD groups. Fecal pellets were freshly collected (8:00 AM of d4, d5, d6 and d13), snap-frozen, and stored at −80°C for later analysis. The animal protocols were carried out in accordance with the Administration System of Laboratory Animals in China and were approved by the Research Ethics Committee of Peking University Sixth Hospital.
16S rRNA Gene Sequencing
The procedures for preparing fecal samples were described previously.14 Briefly, DNA from fecal samples was extracted using the Qiagen QIAmp Fast DNA Stool Mini Kit (Qiagen, Valencia, CA, USA) coupled with an initial bead-beating step as previously described.29 DNA was quantified with a Qubit fluorometer using the Qubit dsDNA BR Assay kit (Invitrogen, Carlsbad, CA, USA). Quality was checked by running an aliquot on 1% agarose gel. Polymerase chain reaction (PCR) enrichment was performed in a 50 μL reaction that contained 30 ng of the template, fusion PCR primer, and PCR master mix. The PCR products were purified with Ampure XP beads and eluted in elution buffer. Total RNA was quantified by spectrofluorimetry and qualified by capillary electrophoresis using Agilent Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA). The validated libraries were used for 2×300 bp paired-end sequencing with the Illumina HiSeq platform (BGI, Shenzhen, China) following the standard pipelines.
Determination of Short-Chain Fatty Acid Concentrations
SCFA levels were determined by gas chromatography-mass spectrometry (GC-MS) in frozen fecal samples that were freshly collected from the rats. The method was adopted from Zhang et al.30 Helium was used as the carrier gas at a constant flow rate of 1 mL/min. The initial oven temperature was held at 60°C for 5 min, increased to 250°C at 10°C/min, and finally held at 250°C for 5 min. The temperatures of the front entrance, transmission line, and electron impact (EI) ion source were set to 280°C, 250°C, and 230°C, respectively. Data processing was performed using an Agilent MSD ChemStation (Agilent). The reported values were normalized to the weight of the original samples that were used.
Statistical Analysis
The statistical analyses were performed using SPSS 20.0 software (IBM, Chicago, IL, USA). The data were assessed for a normal distribution and are expressed as the mean ± SEM. Differences were assessed using repeated-measures analysis of variance (ANOVA) followed by the Bonferroni post hoc test. Values of p < 0.05 were considered statistically significant.
Results
Effects of Sleep Deprivation on Microbial α-Diversity
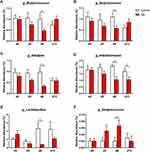
Different diversity indices (ie, Observed species index, Chao1 index, Shannon index, and Simpson index) were used to assess gut microbiota α-diversity (Figure 1B-E). During SD, the mixed-design ANOVA, with group (sleep deprivation and normal sleep control) as the between-groups factor and time of day as the within-groups factor, revealed a main effect of time of day (F3,42 = 9.26, p < 0.001) on the Shannon index of α-diversity and a significant time of day × group interaction (F3,42 = 6.54, p < 0.001; Figure 1D). The ANOVA analysis also showed a significant main effect of time of day (F3,42 = 5.56, p = 0.003) on the Simpson index and a significant time of day × group interaction (F3,42 = 4.95, p = 0.005; Figure 1E). We also found a significant effect of time of day on the Observed species index (F3,42 = 9.23, p < 0.001). The post hoc tests revealed no difference in the α-diversity of the gut microbiota between baseline (Shannon index: p = 0.471; Simpson index: p = 0.318) and after 24 h of SD (Shannon index: p = 0.223; Simpson index: p = 0.329) in SD rats compared with control rats but a significant reduction (Shannon index: p = 0.017; Simpson index: p = 0.021) after 48 h of SD, and this difference disappeared (Shannon index: p = 0.693; Simpson index: p = 0.529) after 1 week of sleep recovery. These results indicate that 48 h of SD reduced α-diversity of the gut microbiota, which fully recovered after 1 week of sleep recovery.
Effects of Sleep Deprivation on Microbial β-Diversity
We further examined the effects of SD on microbial β-diversity, which was performed based on a weighted UniFrac analysis.31 We applied the weighted UniFrac rank (Figure 2A), which provided a measure of the evolutionary distance between microbiotas. We found that the difference between groups was significantly greater than the difference within groups (R = 0.16, p = 0.001). Microbial communities clustered using principal coordinates analysis (PCoA) of the weighted UniFrac matrix. The percentage of variation that was explained by the principal coordinates was indicated on the axes in Figure 2B. PCoA1 represented the main percentage variation (57.82%), revealing significant main effects of time of day (F3,42 = 7.43, p < 0.001) and group (F1,14 = 5.41, p = 0.036) and a significant time of day × group interaction (F3,42 = 2.92, p = 0.045). The PCoA weighted Unifrac matrix analysis showed that the gut microbiome exhibited a trend towards differences (p = 0.068) after 24 h of SD, which became more significant after 48 h of SD (p = 0.004), compared with the normal sleep control group. These differences disappeared (p = 0.597) after 1 week of sleep recovery. These results indicated that 48 h of SD altered β-diversity of the gut microbiota, which fully recovered after 1 week of sleep recovery.
Effects of Sleep Deprivation on Microbial Composition at the Genus Level
To more closely examine the impact of SD on the microbiome, we examined the changes in microbiota composition at the genus level in response to our intervention. The mixed-design ANOVA, with group (SD and normal sleep control) as the between-groups factor and time of day as the within-groups factor, revealed six genera that significantly changed after SD, with a significant time of day × group interaction. There were no significant differences in these altered genera after 24 h of SD (all p > 0.05). After 48 h of SD, the relative abundance of g_Butyricicoccus (p = 0.028), g_Butyricimonas (p = 0.020), g_Alistipes (p = 0.006), g_Intestinimonas (p = 0.001), and g_Lactobacillus (p = 0.014) decreased (Figure 3A-E), whereas the relative abundance of g_Streptococcus (p = 0.040) increased (Figure 3F). After 1 week of sleep recovery, most of the different microbial species recovered such that no differences from the control were found.
Changes in the Stool Content of Short-Chain Fatty Acids Following Sleep Deprivation
Our results showed that the abundance of SCFAs-producing microbes, such as g_Butyricicoccus,32 g_Butyricimonas,33 and g_Alistipes,34 markedly decreased after SD. We further analyzed the levels of total SCFAs and five different SCFAs in samples that were obtained before and after SD and after 1 week of sleep recovery (Figure 4). We found a significant effect of time of day on acetate (F3,42 = 6.00, p = 0.002), propionate (F1.82,25.48 = 12.61, p < 0.001), valerate (F3,42 = 4.73, p = 0.006), and the total amount of fecal SCFAs (F3,42 = 6.27, p = 0.001). We found a significant time of day × group interaction for butyrate (F3,42 = 2.86, p = 0.048), which was significantly reduced after 48 h of SD (p = 0.012). After 1 week of sleep recovery, butyrate concentrations returned to similar levels as controls (p = 0.227; Figure 4D). The correlation analysis showed that fecal butyrate concentrations positively correlated with the relative abundance of g_Butyricimonas (r = 0.39, p = 0.001; Figure 4G). We did not find any interaction for the total amount of SCFAs and other four SCFAs.
Sleep Deprivation Led to Significant Functional Changes in the Gut Microbiota
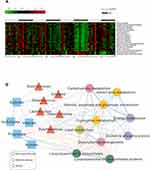
A phylogenetic investigation of communities by the reconstruction of unobserved states (PICRUSt) algorithm was performed to assess the functional differences by plotting differential pathways against the KEGG database with significantly different abundances between SD and control rats.35,36 A total of 637 taxonomies and KEGG ontologies were parsed and mapped into 328 KEGG modules. Compared with the control group, a total of 17 KEGG pathways (energy metabolism, oxidative phosphorylation, carbohydrate metabolism, glycolysis/gluconeogenesis, lipid metabolism, fatty acid metabolism, propanoate metabolism, butanoate metabolism, amino acid metabolism, and tryptophan metabolism, among others) significantly decreased, and two KEGG pathways (lipopolysaccharide biosynthesis and lipopolysaccharide biosynthesis proteins) significantly increased in SD rats (Figure 5A). These differential pathways are mainly involved in four biological processes, especially in various metabolic pathways, such as amino acid metabolism. These results suggested that amino acids, carbohydrates, and lipid metabolism were predominantly disturbed after SD. These functional changes appeared after 24 h of SD and were even more significant after 48 h of SD but with partial restoration after 1 week of sleep recovery. To explore functional correlations between gut microbiota dysbiosis and alterations of fecal metabolites, a correlation matrix was generated using Spearman correlation coefficients between microbial communities at the genus level, SCFAs, and significantly altered KEGG pathways. Overall, the co-correlation analysis showed that KEGG metabolism pathways formed strong and broad co-correlation relationships with both bacterial species and fecal metabolites (Figure 5B).
Discussion
In the present study, we found that total SD led to significant compositional and functional changes in the gut microbiota, and these changes were partially reversible after 1 week of sleep recovery. We examined the impact of 24 h and 48 h of SD and 1 week of sleep recovery on gut microbiota composition in rats. There were no significant differences in gut microbiota composition after 24 h of SD. Significant alterations of gut microbiota composition were observed after 48 h of SD, and this change disappeared after 1 week of sleep recovery. We also found that functional changes in the gut microbiota occurred within 24 h of SD, which were more apparent after 48 h of SD and not fully recovered after 1 week of sleep recovery.
The microbial composition analysis revealed significant alterations of α- and β-diversities after 48 h of SD, indicating that SD may be linked to dynamic changes in intestinal microbiota composition. Similar to the present study, previous studies found that α- and β-diversities of the gut microbiota were significantly altered in patients with insomnia.23,24 A previous study that used an 1-week sleep restriction protocol in both rats and humans found no significant changes in β-diversity or changes in Operational Taxonomic Unit abundance of the gut microbiome.37 A recent study examined a single short period of SD in mice and found no major shifts in gut microbiota composition.26 Consistent with these results, in the present study, no significant difference in gut microbiota composition was found after 24 h of SD. Another study examined the microbiome in humans after 2 days of partial sleep restriction and found that the Firmicutes/Bacteroidetes ratio but not β-diversity was altered by sleep restriction.25 Furthermore, Poroyko et al. found a significant shift in the microbiome in mice after long-term (4 weeks) sleep fragmentation.38 These findings indicated that the microbiome might be resistant to a certain degree of sleep loss, and discrepancies in changes in the gut microbiota might be attributable to the differences in the duration of sleep loss. A short period and slight degree of SD may not be sufficient to exert apparent consequences with regard to dysbiosis of the gut microbiota.
We also identified six significantly altered microbiota at the genus level after 2 days of SD, in which we compared relative abundance between SD and control rats. Sleep-deprived rats exhibited significantly lower proportions of g_Butyricicoccus, g_Butyricimonas, g_Alistipes, g_Intestinimonas, and g_Lactobacillus and significantly higher proportions of g_Streptococcus. Correlation analysis showed that the relative abundance of g_Streptococcus was negatively correlated with the concentration of fecal butyrate and positively correlated with the KEGG pathway of lipopolysaccharide biosynthesis. g_Streptococcus are gram-positive aerobic organisms that cause many disorders, including pharyngitis, pneumonia, wound and skin infections, sepsis, and endocarditis.39 The sleep-wake cycle likewise regulates the normal functioning of the immune system, people have been aware of the sleep loss that accompany many diseases, including multiple microbial infections.40 Sleep loss may enhance the translocation of viable bacteria from the intestine, which provides another means by which sleep–microbe interactions may impact health.41
Total SD is the most common laboratory-based method for studying sleep. Many people often experience SD as a consequence of shift work (eg, truck drivers, physicians, etc.).28,42 Compared with chronic sleep restriction and sleep fragmentation, total SD causes more acute and severe impairment.43 However, the effect of total SD with more than one circadian rhythm on the gut microbiota remains unclear. In previous studies, several methods were used for total SD, such as rotating drums and gentle handling. In the present study, the rats were subjected to 48 h of total SD by gentle handling, a technique of gentle stimulation with physical contact used to disrupt the circadian system and sleep. This method of SD may reduce the animals’ stress.44 Other techniques that have been used for SD in rodents included using a water platform and the presentation of unpleasant stimuli. These techniques, however, are more stressful for the animals than gentle handling.45 Furthermore, environmental factors can contribute to variations in the gut microbiota, leading to inconsistent results.23 To eliminate the influence of the environmental factors and dietary habits on gut microbial communities, we maintained a strict, standard laboratory environment during the experiment.
Evidence suggested that gut bacteria were a source of sleep-inducing signals.46 To determine whether our total SD paradigm functionally affected the intestinal microbiome, we assessed the abundance of fecal SCFAs produced by microbes, such as g_Butyricicoccus,32 g_Butyricimonas,33 and g_Alistipes,34 markedly decreased after SD, which might potentially influence the production of SCFAs. We found a significant effect of time of day on acetate, propionate, valerate, and the total amount of fecal SCFAs. We also found a significant time of day × group interaction for butyrate, in which fecal butyrate significantly decreased after SD compared with the control group. Butyrate is a four-carbon SCFA produced from the fermentation of dietary fibers by microbiota.47 Previous studies found that oral or intraportal butyrate administration robustly increased NREM sleep in rats and induced slow, high-amplitude electroencephalographic wave sleep in rabbits.19,48 Our results were different from previous studies that imposed sleep restriction. In these previous studies, SCFAs did not change after partial SD. This discrepancy in SCFA levels may be attributable to differences in the degree of sleep loss. The present study also found that fecal SCFA content did not change after 24 h of SD.
Sleep deprivation is a biological stressor that alters metabolic parameters. The gut microbiota is now widely accepted to significantly shape several metabolic pathways in the host.12,49 The biological and physiological functions of the gut microbiota can be defined in several ways. Taxonomic composition and diversity are poorly conserved across individuals, whereas genetic composition and functional capacity are evolutionarily conserved across individuals.50 Thus, to decipher the metabolic switch of the gut microbiota, the PICRUSt algorithm was applied to map the bacterial genetic pathway against the KEGG database.23 Compared with the control group, a wide range of pathways were altered in the SD group. These functional changes appeared after 24 h of SD and were even more significant at 48 h of SD. This suggested that some functional changes already appeared during SD before changes in the composition of the gut microbiota were evident. Interestingly, energy metabolism and oxidative phosphorylation pathways significantly decreased in the gut microbiota in SD rats, indicating that SD disturbed energy metabolism.51 Previous study found that SD affected the capacity of mitochondrial bioenergetics, decreasing respiration with the oxidative phosphorylation and electron transport systems.52 The final common metabolic pathway for carbohydrates, lipids, and amino acids is the tricarboxylic acid (TCA) cycle, which is the most effective way for the body to obtain energy through metabolism.53 We found that the carbohydrate, lipid, and amino acid metabolism pathways in the gut microbiota decreased after SD. We also found that glyoxylate, dicarboxylate, and pyruvate metabolism, which belong to the energy and carbohydrate metabolism that were critical for the TCA cycle, were downregulated in the gut microbiota after SD.51,54 Sleep disruption was reported to decrease energy metabolism in rats, suggesting a causal effect of sleep loss on weight gain.55,56 Moreover, lipopolysaccharide biosynthesis pathways also increased in the gut microbiota in SD rats. Lipopolysaccharide causes an inflammatory reaction mainly via activation of the Toll-like receptor 4/nuclear factor κB inflammatory pathways. Human and rodent studies found that sleep loss increased peripheral markers of inflammation.57–59 Chronic sleep loss can alter the gut microbiota and induce systemic inflammation in mice.38 Disturbances in the metabolism of amino acid neurotransmitters, such as tryptophan, glutamate, and γ-aminobutyric acid (GABA), are prominent after SD. Recent studies showed that the gut microbiome might indicate the onset of mental disorders by modulating amino acid neurotransmitters, such as GABA and serotonin. As a precursor for production of the neurotransmitter serotonin, tryptophan metabolism has also been found to be altered by SD and linked to various neurodegenerative diseases.60,61 In the present study, we found that the levels of fecal amino acid neurotransmitters (ie, glutamate, tryptophan, aspartate, and glycine) decreased in SD rats, suggesting a decrease in amino acid neurotransmitter content in the intestine after SD. These findings suggested that fecal amino acid metabolism might be modulated by gut microbes, which might reflect detrimental consequences of sleep loss.
There are some limitations of the current study. First, this study used 16S rRNA gene sequencing analysis without further metagenomic analysis. Although more data and information can be obtained through the shotgun metagenomics, 16S rRNA gene sequencing is currently more commonly used and popular in microbiome research.62 Second, unique perturbations to the ileal microbiota that register even after acute SD suggest that analysis of fecal material alone may not be appropriate when assessing dysbiosis of the gut microbiome.20 Third, we have only conducted correlation analysis for gut microbiota and metabolism pathways, which could not provide evidence of causality. We also recognize that the sample size in each group was limited. We plan to explore the mechanism using large sample sizes in future studies.
Conclusion
In the present study, we examined the impact of 24 and 48 h of total SD and 1 week of sleep recovery on gut microbiota composition and function. We found that 48 h but not 24 h of total SD significantly altered composition of the gut microbiota, which were reversible after 1 week of sleep recovery. We also found that functional changes in the gut microbiota occurred within 24 h of SD, which was more apparent after 48 h of SD and did not fully recover after 1 week of sleep recovery. These findings indicated that SD might be linked to dynamic changes in composition and function of the intestinal microbiota. Future studies should elucidate the molecular mechanisms that are responsible for these changes and how they contribute to behavioral and neurophysiological phenotypes that are associated with sleep loss.
Acknowledgments
This work was supported in part by the National Key Research and Development Program of China (no. 2020YFC2003600), National Natural Science Foundation of China (no. 81901347, 82171477, 31800897, 81821092, and 82001404), Peking University Medicine Fund for Fostering Young Scholars’ Scientific and Technological Innovation, and Fundamental Research Funds for Central Universities (no. BMU2018PYB010). We thank Shi-Qiu Meng and Ya-Bin Wei for their help in revising our manuscript. We are grateful to BioNovoGene (Suzhou) for assistance with GC-MS.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Nagari M, Gera A, Jonsson S, Bloch G. Burmble bee workers give up sleep to care for offspring that are not their own. Curr Biol. 2019;29(20):3488–3493. doi:10.1016/j.cub.2019.07.091
2. Salfi F, Lauriola M, Tempesta D, et al. Effects of total and partial sleep deprivation on reflection impulsivity and risk-taking in deliberative decision-making. Nat Sci Sleep. 2020;12:309–324. doi:10.2147/nss.S250586
3. Slavish DC, Taylor DJ, Lichstein KL. Intraindividual variability in sleep and comorbid medical and mental health conditions. Sleep. 2019;42:6. doi:10.1093/sleep/zsz052
4. Reynolds AC, Paterson JL, Ferguson SA, Stanley D, Wright KP
5. Li JQ, Wang YH, Lu QD, Xu YY, Bao YP. Prevalence of psychological distress in type ii diabetes in China: a systematic review and meta-analysis. Heart Mind. 2019;3(4):147. doi:10.4103/hm.hm_67_19
6. Sun Y, Bao Y, Ravindran A, Sun Y, Lu L. Mental health challenges raised by rapid socioeconomic transformations in China: lessons learned and prevention strategies. Heart Mind. 2020;4(3):59. doi:10.4103/hm.hm_22_20
7. Liu R, Hong J, Xu X, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23(7):859–868. doi:10.1038/nm.4358
8. Lee CJ, Sears CL, Maruthur N. Gut microbiome and its role in obesity and insulin resistance. Ann N Y Acad Sci. 2020;1461(1):37–52. doi:10.1111/nyas.14107
9. Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. doi:10.1038/nature15766
10. Morais LH, Schreiber H, Mazmanian SK. The gut microbiota-brain axis in behaviour and brain disorders. Nat Rev Microbiol. 2020;19(4):241–255. doi:10.1038/s41579-020-00460-0
11. Sgritta M, Dooling SW, Buffington SA, et al. Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron. 2019;101(2):246–259 e246. doi:10.1016/j.neuron.2018.11.018
12. Zhu F, Guo R, Wang W, et al. Transplantation of microbiota from drug-free patients with schizophrenia causes schizophrenia-like abnormal behaviors and dysregulated kynurenine metabolism in mice. Mol Psychiatry. 2019;25(11):2905–2918. doi:10.1038/s41380-019-0475-4
13. Pearson-Leary J, Zhao C, Bittinger K, et al. The gut microbiome regulates the increases in depressive-type behaviors and in inflammatory processes in the ventral hippocampus of stress vulnerable rats. Mol Psychiatry. 2019;25(5):1068–1079. doi:10.1038/s41380-019-0380-x
14. Zheng P, Zeng B, Zhou C, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21(6):786–796. doi:10.1038/mp.2016.44
15. Wong ML, Inserra A, Lewis MD, et al. Inflammasome signaling affects anxiety- and depressive-like behavior and gut microbiome composition. Mol Psychiatry. 2016;21(6):797–805. doi:10.1038/mp.2016.46
16. Saad MJ, Santos A, Prada PO. Linking gut microbiota and inflammation to obesity and insulin resistance. Physiology (Bethesda). 2016;31(4):283–293. doi:10.1152/physiol.00041.2015
17. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi:10.1016/j.cell.2016.05.041
18. Sivaprakasam S, Prasad PD, Singh N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol Ther. 2016;164:144–151. doi:10.1016/j.pharmthera.2016.04.007
19. Szentirmai É, Millican NS, Massie AR, Kapás L. Butyrate, a metabolite of intestinal bacteria, enhances sleep. Sci Rep. 2019;9(1):7035. doi:10.1038/s41598-019-43502-1
20. Triplett J, Ellis D, Braddock A, et al. Temporal and region-specific effects of sleep fragmentation on gut microbiota and intestinal morphology in Sprague Dawley rats. Gut Microbes. 2020;11(4):1–15. doi:10.1080/19490976.2019.1701352
21. Matenchuk BA, Mandhane PJ, Kozyrskyj AL. Sleep, circadian rhythm, and gut microbiota. Sleep Med Rev. 2020;53:101340. doi:10.1016/j.smrv.2020.101340
22. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi:10.1038/4441022a
23. Liu B, Lin W, Chen S, et al. Gut microbiota as a subjective measurement for auxiliary diagnosis of insomnia disorder. Front Microbiol. 2019;10:1770. doi:10.3389/fmicb.2019.01770
24. Li Y, Zhang B, Zhou Y, et al. Gut microbiota changes and their relationship with inflammation in patients with acute and chronic insomnia. Nat Sci Sleep. 2020;12:895–905. doi:10.2147/nss.S271927
25. Benedict C, Vogel H, Jonas W, et al. Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Mol Metabol. 2016;5(12):1175–1186. doi:10.1016/j.molmet.2016.10.003
26. El Aidy S, Bolsius YG, Raven F, Havekes R. A brief period of sleep deprivation leads to subtle changes in mouse gut microbiota. J Sleep Res. 2020;29(6):e12920. doi:10.1111/jsr.12920
27. Oonk M, Davis CJ, Krueger JM, Wisor JP, Van Dongen HP. Sleep deprivation and time-on-task performance decrement in the rat psychomotor vigilance task. Sleep. 2015;38(3):445–451. doi:10.5665/sleep.4506
28. Shi HS, Luo YX, Xue YX, et al. Effects of sleep deprivation on retrieval and reconsolidation of morphine reward memory in rats. Pharmacol Biochem Behav. 2011;98(2):299–303. doi:10.1016/j.pbb.2011.01.006
29. Bruce-Keller AJ, Salbaum JM, Luo M, et al. Obese-type gut microbiota induce neurobehavioral changes in the absence of obesity. Biol Psychiatry. 2015;77(7):607–615. doi:10.1016/j.biopsych.2014.07.012
30. Zhang L, Wang Y, Xiayu X, et al. Altered gut microbiota in a mouse model of Alzheimer’s disease. J Alzheimers Dis. 2017;60(4):1241–1257. doi:10.3233/jad-170020
31. Li N, Wang Q, Wang Y, et al. Oal probiotics ameliorate the behavioral deficits induced by chronic mild stress in mice via the gut microbiota-inflammation axis. Front Behav Neurosci. 2018;12:266. doi:10.3389/fnbeh.2018.00266
32. Chang SC, Shen MH, Liu CY, Pu CM, Hu JM, Huang CJ. A gut butyrate-producing bacterium Butyricicoccus pullicaecorum regulates short-chain fatty acid transporter and receptor to reduce the progression of 1,2-dimethylhydrazine-associated colorectal cancer. Oncol Lett. 2020;20(6):327. doi:10.3892/ol.2020.12190
33. Duan C, Kuang L, Xiang X, et al. Activated Drp1-mediated mitochondrial ROS influence the gut microbiome and intestinal barrier after hemorrhagic shock. Aging (Albany NY). 2020;12(2):1397–1416. doi:10.18632/aging.102690
34. McKenna A, Ijaz UZ, Kelly C, et al. Impact of industrial production system parameters on chicken microbiomes: mechanisms to improve performance and reduce Campylobacter. Microbiome. 2020;8(1):128. doi:10.1186/s40168-020-00908-8
35. Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi:10.1038/nbt.2676
36. Zhou X, Liu L, Lan X, et al. Polyunsaturated fatty acids metabolism, purine metabolism and inosine as potential independent diagnostic biomarkers for major depressive disorder in children and adolescents. Mol Psychiatry. 2019;24(10):1478–1488. doi:10.1038/s41380-018-0047-z
37. Zhang SL, Bai L, Goel N, et al. Human and rat gut microbiome composition is maintained following sleep restriction. Proc Natl Acad Sci U S A. 2017;114(8):E1564–E1571. doi:10.1073/pnas.1620673114
38. Poroyko VA, Carreras A, Khalyfa A, et al. Chronic sleep disruption alters gut microbiota, induces systemic and adipose tissue inflammation and insulin resistance in mice. Sci Rep. 2016;6:35405. doi:10.1038/srep35405
39. Baracco GJ. Infections caused by Group C and G Streptococcus (Streptococcus dysgalactiae subsp. equisimilis and Others): epidemiological and clinical aspects. Microbiol Spectr. 2019;7(2). doi:10.1128/microbiolspec.GPP3-0016-2018
40. Krueger JM, Toth LA, Floyd R, et al. Sleep, microbes and cytokines. Neuroimmunomodulation. 1994;1(2):100–109. doi:10.1159/000097142
41. Krueger JM, Opp MR. Sleep and microbes. Int Rev Neurobiol. 2016;131:207–225. doi:10.1016/bs.irn.2016.07.003
42. Chen WH, Chen J, Lin X, et al. Dissociable effects of sleep deprivation on functional connectivity in the dorsal and ventral default mode networks. Sleep Med. 2018;50:137–144. doi:10.1016/j.sleep.2018.05.040
43. Reynolds AC, Banks S. Total sleep deprivation, chronic sleep restriction and sleep disruption. Prog Brain Res. 2010;185:91–103. doi:10.1016/B978-0-444-53702-7.00006-3
44. Lemons A, Saré RM, Beebe Smith C. Chronic sleep deprivation in mouse pups by means of gentle handling. J Vis Exp. 2018;140. doi:10.3791/58150
45. Blumberg MS, Middlemis-Brown JE, Johnson ED. Sleep homeostasis in infant rats. Behav Neurosci. 2004;118(6):1253–1261. doi:10.1037/0735-7044.118.6.1253
46. Ogawa Y, Miyoshi C, Obana N, et al. Gut microbiota depletion by chronic antibiotic treatment alters the sleep/wake architecture and sleep EEG power spectra in mice. Sci Rep. 2020;10(1):19554. doi:10.1038/s41598-020-76562-9
47. Matt SM, Allen JM, Lawson MA, Mailing LJ, Woods JA, Johnson RW. Butyrate and dietary soluble fiber improve neuroinflammation associated with aging in mice. Front Immunol. 2018;9:1832. doi:10.3389/fimmu.2018.01832
48. Samson FE
49. Yang J, Zheng P, Li Y, et al. Landscapes of bacterial and metabolic signatures and their interaction in major depressive disorders. Sci Adv. 2020;6:49. doi:10.1126/sciadv.aba8555
50. Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23(3):314–326. doi:10.1038/nm.4272
51. Ma W, Song J, Wang H, et al. Chronic paradoxical sleep deprivation-induced depression-like behavior, energy metabolism and microbial changes in rats. Life Sci. 2019;225:88–97. doi:10.1016/j.lfs.2019.04.006
52. Rodrigues NR, Macedo GE, Martins IK, et al. Short-term sleep deprivation with exposure to nocturnal light alters mitochondrial bioenergetics in Drosophila. Free Radic Biol Med. 2018;120:395–406. doi:10.1016/j.freeradbiomed.2018.04.549
53. Akram M. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem Biophys. 2014;68(3):475–478. doi:10.1007/s12013-013-9750-1
54. Liu X, Yang Y, Zhao M, et al. Proteomics reveal energy metabolism and mitogen-activated protein kinase signal transduction perturbation in human Borna disease virus Hu-H1-infected oligodendroglial cells. Neuroscience. 2014;268:284–296. doi:10.1016/j.neuroscience.2014.03.009
55. St-Onge MP. Sleep-obesity relation: underlying mechanisms and consequences for treatment. Obes Rev. 2017;18(Suppl 1):34–39. doi:10.1111/obr.12499
56. Coborn JE, Lessie RE, Sinton CM, Rance NE, Perez-Leighton CE, Teske JA. Noise-induced sleep disruption increases weight gain and decreases energy metabolism in female rats. Int J Obes (Lond). 2019;43(9):1759–1768. doi:10.1038/s41366-018-0293-9
57. Irwin MR. Sleep and inflammation: partners in sickness and in health. Nat Rev Immunol. 2019;19(11):702–715. doi:10.1038/s41577-019-0190-z
58. Irwin MR, Wang M, Ribeiro D, et al. Sleep loss activates cellular inflammatory signaling. Biol Psychiatry. 2008;64(6):538–540. doi:10.1016/j.biopsych.2008.05.004
59. Wang Z, Chen WH, Li SX, et al. Gut microbiota modulates the inflammatory response and cognitive impairment induced by sleep deprivation. Mol Psychiatry. 2021;26(11):6277–6292. doi:10.1038/s41380-021-01113-1
60. Bhat A, Pires AS, Tan V, Babu Chidambaram S, Guillemin GJ. Effects of sleep deprivation on the tryptophan metabolism. Int J Tryptophan Res. 2020;13:1178646920970902. doi:10.1177/1178646920970902
61. Strasser B, Gostner JM, Fuchs D. Mood, food, and cognition: role of tryptophan and serotonin. Curr Opin Clin Nutr Metab Care. 2016;19(1):55–61. doi:10.1097/mco.0000000000000237
62. Schriefer AE, Cliften PF, Hibberd MC, et al. A multi-amplicon 16S rRNA sequencing and analysis method for improved taxonomic profiling of bacterial communities. J Microbiol Methods. 2018;154:6–13. doi:10.1016/j.mimet.2018.09.019
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.