Back to Journals » Journal of Inflammation Research » Volume 14
Alterations in the Plasma Proteome Induced by SARS-CoV-2 and MERS-CoV Reveal Biomarkers for Disease Outcomes for COVID-19 Patients
Authors Alaiya A , Alshukairi A, Shinwari Z, AL-Fares M, Alotaibi J, AlOmaim W, Alsharif I, Bakheet R, Alharbi L, Allam R, Asiri A, Memish Z , Alromaih K, Al-Mozaini M
Received 1 June 2021
Accepted for publication 28 July 2021
Published 1 September 2021 Volume 2021:14 Pages 4313—4328
DOI https://doi.org/10.2147/JIR.S322430
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Monika Sharma
Ayodele Alaiya,1 Abeer Alshukairi,2 Zakia Shinwari,1 Mariam AL-Fares,3 Jawaher Alotaibi,4 Waleed AlOmaim,3 Ibtihaj Alsharif,5 Razan Bakheet,6 Layla Alharbi,5 Rabab Allam,1 Ayed Asiri,7 Ziad Memish,8 Khaldoun Alromaih,6 Maha Al-Mozaini5
1Proteomics Unit, Stem Cell and Tissue Re-Engineering Program, King Faisal Specialist Hospital and Research Centre, Riyadh, 11211, Saudi Arabia; 2Department of Medicine, King Faisal Specialist Hospital and Research Center, Jeddah, Kingdom of Saudi Arabia; 3Clinical Chemistry Laboratory, Department of Pathology and Laboratory Medicine, King Faisal Specialist Hospital and Research Centre, Riyadh, Saudi Arabia; 4Department of Medicine, King Faisal Specialist Hospital and Research Center, Riyadh, Kingdom of Saudi Arabia; 5Immunocompromised Host Research Unit, Department of Infection and Immunity, King Faisal Specialist Hospital and Research Centre, Riyadh, Saudi Arabia; 6Centre for Genomic Medicine, King Faisal Specialist Hospital and Research Center, Riyadh, Kingdom of Saudi Arabia; 7Critical Care Services, Al Imam Abdulrahman Al Faisal Hospital, Riyadh, Kingdom of Saudi Arabia; 8Research and Innovation Center, King Saud Medical City, Ministry of Health, Riyadh, Kingdom of Saudi Arabia
Correspondence: Ayodele Alaiya
Proteomics Unit, Stem Cell and Tissue Re-Engineering Program, King Faisal Specialist Hospital and Research Centre, KFSH&RC, BOX 3354, Riyadh, 11211, Saudi Arabia
Tel +966 114424178
Email [email protected]
Maha Al-Mozaini
Immunocompromised Host Research Unit, Department of Infection and Immunity, KFSH&RC, Riyadh, Saudi Arabia
Tel +966 114439632
Email [email protected]
Purpose: This study aimed to understand the pathophysiology of host responses to infections caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)/(COVID-19) and Middle East respiratory syndrome coronavirus (MERS-CoV) and to identify proteins for patient stratification with different grades of illness severity.
Patients and Methods: Peripheral blood samples from 43 patients with different grades of COVID-19, 7 MERS-CoV patients admitted to the ICU, and 10 healthy subjects were analyzed using label-free quantitative liquid chromatography–mass spectrometry (LC–MS).
Results: We identified 193 and 91 proteins that differed significantly between COVID-19 and MERS-CoV sample groups, respectively, and 49 overlapped between datasets. Only 10 proteins are diagnostic of asymptomatic cases, 12 are prognostic of recovery from severe illness, and 28 are prognostic of a fatal outcome of COVID-19. These proteins are implicated in virus-specific/related signaling networks. Notable among the top canonical pathways are humoral immunity, inflammation, acute-phase response signaling, liver X receptor/retinoid X receptor (LXR/RXR) activation, coagulation, and the complement system. Furthermore, we confirmed positive viral shedding in 11.76% of 51 additional peripheral blood samples, indicating that caution should be taken to avoid the possible risk of transfusion of infected blood products.
Conclusion: We identified COVID-19 and MERS-CoV protein panels that have potential as biomarkers and might assist in the prognosis of SARS-CoV-2 infection. The identified markers further our understanding of COVID-19 disease pathophysiology and may have prognostic or therapeutic potential in predicting or managing host cell responses to human COVID-19 and MERS-CoV infections.
Keywords: COVID-19, SARS-CoV, MERS-CoV, expression proteomics, biomarkers
Brief Commentary
Background
Identifying key protein–protein interactions involved in responses to different coronavirus infections may improve disease management. We analyzed the peripheral blood of patients with different severity grades of COVID-19 and MERS-CoV to determine whether there were distinct molecular changes characteristic of disease outcomes.
Translational Significance
We identified objective markers that may be used to predict COVID-19 outcomes at the point of care and biomarkers to identify asymptomatic individuals; those who will progress to develop mild, moderate, or severe disease; and those who will recover without life-threatening complications of COVID-19. This study supports some previous findings and describes new additional diagnostic markers for asymptomatic individuals. The results also offer a means of objectively predicting disease outcomes among severely ill COVID-19 patients.
Introduction
Within the past two decades, there have been three major coronavirus outbreaks: the first two were SARS-CoV-1 in 2002 and the Middle East respiratory syndrome coronavirus (MERS-CoV) epidemic outbreak in the Arabian Peninsula in 2012.1,2 MERS-CoV infected about 3000 individuals, causing approximately 850 deaths. The most recent outbreak of a new coronavirus, SARS-CoV-2, was first reported in Wuhan, China, and causes the disease known as COVID-19.3–7 The outbreak developed into a pandemic, and the latest report from the World Health Organization (WHO) as of March 1st, 2021, indicates that well over 123 million people have been infected, with more than 2.7 million deaths. All coronaviruses belong to a large family of viruses known to cause respiratory illness, with clinical manifestations including but not limited to lung infections, flu-like symptoms, and severe acute respiratory distress.
Presently, reports show that SARS-CoV-2 is less pathogenic than SARS-CoV-1 and MERS viruses but poses an unprecedented global threat due to its high transmission rate.8 Although a large fraction of infected people are asymptomatic, the disease is aggressively virulent in some patients, many of whom progress to hospitalization and require Intensive Care Unit (ICU) admission. Worldwide, the overall mortality rate is 2–15%, which varies depending on age and other underlying health problems.9–11
As the COVID-19 pandemic is still ongoing, comparative analysis with previous coronavirus-related diseases such as MERS-CoV may be particularly informative. Different gene expression studies, including those applying proteomic approaches, have been performed using biological samples from COVID-19 patients.12–14 Many of these studies have focused primarily on COVID-19. However, inter-laboratory comparative analysis of the generated data has been challenging because of differences in the technologies used, pre-analysis sample handling, and lack of a standardized universal protocol.15 In the present study, the peripheral blood plasma of patients with different severity grades of COVID-19 and MERS-CoV were analyzed to determine whether there were distinct molecular changes characteristic of disease outcomes. We anticipate that these molecular changes will improve our understanding of the pathophysiology of the disease and aid in the discovery of therapeutic targets for both SARS-CoV-2 and MERS-CoV patients.
The added value and uniqueness of this study lie in the discovery of objective markers that may be used to predict COVID-19 outcomes at the point of care and to identify asymptomatic individuals, those who will progress to develop mild, moderate, or severe disease, and those who will recover without life-threatening complications of COVID-19.
Materials and Methods
Study Design and Subjects
This study included three different groups: (1) One hundred and twenty-two confirmed COVID-19 cases, comprising individuals who were asymptomatic and those with mild, moderate, and severe infections with respiratory tract symptoms as well as multi-organ involvement; (2) seven cases of severely infected individuals with MERS-CoV, all of whom were admitted to the ICU; and (3) ten negative control subjects with flu-like symptoms who were RT-qPCR negative for both SARS-CoV-2 and MERS-CoV. All COVID-19-positive cases were admitted to King Faisal Specialist Hospital and Research Center, Riyadh (KFSH&RC-Riyadh), Saudi Arabia, between March and September 2020, while patients with MERS-CoV were admitted between 2018 and 2019. Our institutional Office of Research Affairs and Internal Review Board at KFSHRC-Riyadh approved the study (MERS: RAC# 2180024 and COVID-19 RAC# 2200011). The clinical and demographic characteristics of all analyzed samples are detailed in Table 1, and the analysis workflow for this study is illustrated in Figure 1.
 |
Table 1 Clinical and Demographic Characteristics of Analyzed COVID-19 and MERS- CoV Infected Patients’ samples |
 |
Figure 1 Workflow illustration. Peripheral blood samples collected from different sample cohorts were pre-processed. The biomaterials were inactivated prior to proteomic analysis, and complex differentially expressed protein (DEP) data were subjected to multivariate computational analysis for biological interpretation of the different stages of the coronavirus disease process. The datasets were filtered, and potential COVID-19-related biomarker panels were identified. The details of the clinical and demographic characteristics of all the samples are listed in.Table 1 |
Sample Collection and Preparation Prior to Protein Expression Analysis
Peripheral blood plasma (PBP) samples from patients with an RT-qPCR-confirmed diagnosis of SARS-CoV-2 or MERS-CoV were separated into plasma and serum for proteomic analysis and viral load measurements, respectively. Prior to proteomic analysis, the PBP samples were depleted of common high-abundance plasma proteins using human albumin removal reagents from Agilent Technologies (Santa Clara, California, United States). Label-free quantitative expression proteomics by liquid chromatography–tandem mass spectrometry (LC–MS/MS) was used for the analysis of the complex protein mixtures from all sample groups.
Biological and Analytical Replicates
Because of the inherently low throughput of the LC–MS/MS analysis platform, only samples with the same diagnosis and with comparable values of total protein contents, as assessed by protein concentration assay, were pooled together into an analysis cohort. This practice in LC–MS-based analysis has potential risks of confounding the results if due care is not taken.16,17 All samples within an analysis group were initially pre-screened for intra-sample homogeneity prior to pooling as previously described.16
All other samples that were excluded in the expression analysis were evaluated in the validation phase of the analysis for variability in individual changes observed in the analyzed pooled samples. Each group of pooled samples was run in triplicate to measure method reproducibility and minimize instances of missing values in the generated datasets.
Handling COVID-19 Biomaterial, Biosafety, and Pathogen Inactivation
The currently known coronaviruses are usually transmitted by direct contact with infectious pathogens via the upper or lower respiratory tract rather than through the blood-borne route. However, viral shedding in bodily fluids such as plasma or serum has been reported to vary between 1 and 3%.18–21 Different pathogen inactivation methods for coronaviruses have been described, including different solvents and detergents as well as heat inactivation.18 We used a modified heat inactivation method prior to handling the biomaterials, as previously reported.18 Briefly, all peripheral blood samples were heat-inactivated at 65 °C for 30 min, and all samples were handled in accordance with BSC IIA protocols. The experimental setup and analysis workflow for this study are illustrated in Figure 1.
Data Collection
Collaborating physicians and research staff collected clinical data from hospital electronic medical records. The clinical data were then analyzed with respect to SARS-CoV-2 viral dynamics, including demographics and comorbidities. The documented comorbidities included diabetes mellitus, heart disease, chronic lung disease, renal failure, and other conditions such as cancer (Table 1). We considered the symptoms to have started when any of the following appeared: fever, cough, chills, dizziness, headache, and fatigue. The severity of illness was evaluated according to guidelines for the diagnosis and management of coronavirus disease 2019 (COVID-19) updated in late November 2020 by WHO. The modified descriptions of the grade and clinical spectrum of analyzed COVID-19 samples are summarized in Table 1.
Protein Characterization by Mass Spectrometry Analysis
In-Solution Protein Digestion
Crude plasma from each patient was first depleted of the top ten most abundant plasma proteins using a human albumin removal kit (Agilent Technologies), and the protein concentrations of all samples were normalized as previously described.16 A total of 100 µg comprising equal amounts of protein contributed by each sample constituting a pool of one analysis cohort was in-solution digested prior to LC–MS/MS analysis, as previously described.16 Briefly, complex proteins were heat-denatured at 80 °C for 15 min, reduced in 10 mM DTT at 60 °C for 30 min, and then alkylated in 10 mM iodoacetamide (IAA) for 40 min at room temperature in the dark. The calculated amount of 1 µg/µL trypsin (Promega, US) was added (50:1 sample/trypsin ratio) for overnight digestion at 37 °C. All samples were normalized-diluted with aqueous 0.1% formic acid to a concentration of 1 µg/µL, and 3 µL (corresponding to 3 µg) of each sample was injected for LC–MS/MS analysis. All the samples were spiked with yeast alcohol dehydrogenase (ADH; P00330) as an internal standard for quantitation.
Protein Identification
The resulting peptides from the in-solution-digested proteins were analyzed using one-dimensional Nano Acquity liquid chromatography coupled to Synapt G2 HDMS on a Trizaic Nano-flow source (Waters, Manchester, UK). Complex proteomic data were generated between the sample groups as previously described.16,22 The MS data were acquired in an m/z range of 50–2000 Da with a gradient acquisition run time of 120 min using ion-mobility separation experiments (HDMSE). Each sample was analyzed in triplicate runs using the Mass Lynx platform (Version. 4·1, SCN833).
Bioinformatics and Data Analysis
Progenesis QI for proteomics (QIfP) V 3.0 (Waters/Nonlinear Dynamics, UK) was used for all automated data processing and database searching, and differentially expressed proteins between sample groups were identified as previously described.16,23 Briefly, the acquired list of peptide ions for protein identification was queried in the unified UniProt/SwissProt non-redundant human protein sequence database (www.uniprot.org). The list of identified protein datasets was further reviewed using multivariate statistical analyses. Normalized label-free quantification was performed for each of the identified proteins using the ‘Hi3ʹ algorithm in Progenesis QI software (Nonlinear Dynamics/Waters). A known protein, alcohol dehydrogenase (ADH, P00330), was used as an internal standard to determine the absolute amount of each identified protein as previously described.16,23 The generated expression data were filtered using two overlapping parameters to include only proteins that were markedly statistically different (ANOVA, P ≤ 0.05; >2-fold change in expression) and the so-called on/off proteins (present or absent). The derived differentially expressed protein datasets were subjected to principal component analysis (PCA) and hierarchical cluster analysis.
COVID-19 Viral Detection in Serum/Plasma Samples and Multiplex One-Step RT-qPCR SARS-CoV-2 Assay
Serum samples were stored at −80 °C until viral RNA extraction in the BSL3 research laboratory. Five-hundred-microliter aliquots were used for each extraction. Viral RNA extraction was conducted by our in-house automated RNA extraction protocol using KingFisher FlexTM from Thermo Fisher Scientific, as previously described in.24 Samples with cycle threshold (CT) values of ≤40.0 were considered positive for SARS-CoV-2 RNA. Viral load was calculated by plotting CT values against the SARS-CoV-2 RNA template.
Results
Development of Protein Biomarker Panel for Prediction of Disease Progression from Asymptomatic to Mild, Moderate, and Severe COVID-19 Infection
Overall, we evaluated 122 peripheral blood samples from patients with different grades of COVID-19, including asymptomatic, mild, moderate, and severe cases. Owing to the low throughput of the mass spectrometry analysis platform, only 43 samples were subjected to quantitative proteomics analysis. Fifty-one (51) samples were analyzed for viral shedding, and 28 samples were not sufficient for proteomic analysis. The experimental setup and analysis workflow for this study are illustrated in Figure 1. The mean age of all patients was 47 years (range 02–86 years). The clinical and demographic characteristics of all analyzed samples are detailed in Table 1. All but severe cases of COVID-19 recovered from the disease, and 6 of 12 severely ill patients admitted to the ICU died of the disease. The average number of identified proteins from triplicate runs of each sample group was 303, of which 193 were significantly differentially expressed (≥2-fold change and ANOVA p < 0.05) between all five sample groups (Supplementary Table S1). We observed distinct sample clusters using unsupervised principal component analysis and hierarchical cluster analysis of the 193 proteins. Asymptomatic, mild, and moderate cases of the disease formed a cluster that was distinct from ICU-admitted patients who recovered from severe illness and ICU-admitted patients who died of the disease (Figure 2A). A similar pattern was observed in the heat map of comparative protein expression patterns of asymptomatic, mild, and moderate cases against the profiles of severe patients (Figure 2A).
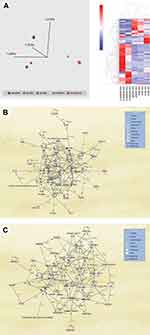 |
Figure 2 (A) (Left panel) Principal component analysis (PCA) using the expression dataset of 193 differentially expressed proteins between grades of COVID-19 patients. Three distinct clusters are apparent, with asymptomatic (Asymp), mild, and moderate (Mod) forming a close cluster distinct from ICU-admitted patients who recovered from severe illness (Severe-R) and ICU-admitted patients who died of severe illness (Severe-D). (Right panel) Hierarchical cluster analysis using the above dataset of the 193 differentially expressed proteins between grades of COVID-19 patients. Asymptomatic, mild, and moderate cases show remarkably similar heat map patterns, while recovered severely ill and deceased severely ill are distinctively different. The heat map shows the relative amounts of proteins by color as either upregulation with positive z-scores in red or downregulation with negative z-scores in blue. The representations of the trend in the expression changes of some of these proteins between all five grades of COVID-19 patients are illustrated in Supplementary Figure 1, and a complete list of the proteins is in Supplementary Table S1. The figure was generated using Qlucore Omics Explorer version 3.7 (Lund, Sweden). (B) Ingenuity pathway analysis of the 193 differentially expressed proteins between the five grades of COVID-19. Some of the 193 proteins that were mapped/implicated in the IPA database in the pathway analysis of two signaling networks, including Cell Death and Survival, Cardiovascular Disease, and Cell-To-Cell Signaling and Interaction. (C) Cellular Compromise, Inflammatory Response, and Hematological System Development and Function. (The figure was partly generated using a licensed Ingenuity Pathway Analysis program (www.qiagen.com)). |
Analysis of Cellular Processes and Biofunctional Annotations of Identified Proteins
The 193 identified differentially expressed proteins between the five grades of COVID-19 patients were further assessed for their functional characteristics and implications in infectious disease and immune-mediated disorders using Ingenuity Pathway Analysis (IPA). The majority of the 193 proteins were associated with four signaling networks: (1) cellular function and maintenance, metabolic disease, and organismal injury and abnormalities; (2) humoral immune response, inflammatory response, and hematological system development and function; (3) cell death and survival, cardiovascular disease, and cell-to-cell signaling and interaction; (4) molecular transport, cellular assembly and organization, and cellular compromise.
The biofunctional annotations of these proteins include, among others, calcium channel, transmembrane receptor, activator, and enzyme, while other functions include transporter, peptidase, and kinase.
The protein–protein interactions and gene names of these proteins are highlighted in Figure 2B and C. The representations of expression changes of some of the differentially expressed proteins between samples from SARS-CoV-2-infected patients with five different grades of severity are illustrated in Supplementary Figure 1.
Canonical/Essential Pathway Analysis and Pathophysiology of COVID-19
The associations and protein–protein interactions of the molecules were further evaluated to assess whether they were directly involved in virus-specific/related signaling networks. Notable among the top canonical/essential pathways are acute-phase response signaling, liver X receptor/retinoid X receptor (LXR/RXR) activation, Farnesoid X receptor (FXR)/RXR activation, complement system, and atherosclerosis signaling, with several implicated molecules overlapping between the different pathways. The interactions of some of the proteins with respect to their involvement in the pathophysiology of COVID-19 are summarized in Figure 3A. Comparison of the canonical pathways and toxicity functions in different pairwise analyses of COVID-19 patients showed higher gene expression changes (overexpression) between severe and asymptomatic COVID-19 pairs than between mild or moderate and asymptomatic disease pairs (Supplementary Figure 2). We observed that proteins associated with LXR/RXR activation, acute-phase reaction, and production of nitric oxide and reactive oxygen species were upregulated in severely ill COVID-19 patients compared with the other groups with less severe disease. The expression changes of the implicated molecules were distinct, as shown in the heat maps in Figure 3B.
The cellular process and disease annotations of some of these protein expression signatures among the different grades of SARS-CoV-2 infections are described in Supplementary Table S1 and Supplementary Figure 3A–C.
Viral Shedding in Blood of COVID-19 Patients
Previous reports have suggested an association between viral disease severity and shedding in biological samples.18–21 Blood samples with adequate amounts of RNA from the SARS-CoV-2 cohort were subjected to viral RNA extraction followed by RT-qPCR to identify SARS-CoV-2 genes. We were able to conduct this experiment on 51 samples (41% of the total number). We detected positive viral shedding in 6 (11.76%) of the 51 samples (Table 1). Three of the six samples were collected from patients who were diagnosed with grade D (severe) disease, and the other three samples were from patients who progressed from grade C (moderate) to grade D (severe).
Protein Signatures for Disease Outcomes of ICU-Admitted MERS-CoV Patients for the Discovery of Predictive Markers
We analyzed PBP sample groups of severe MERS-CoV patients (n = 7) who required ICU admission and died of the disease and those who survived the infection. Samples of healthy subjects without a history of MERS-CoV infection and seasonal flu infection were used as the negative control group (n = 10). The mean age of all ICU patients who survived is 57 years (range 31–75 years), while the mean age of all ICU patients who died is 45.6 years (range 42–49 years). Table 1 details the clinical and demographic characteristics of all analyzed samples. The average number of proteins identified in triplicate runs of the samples was 235, of which only 91 proteins were significantly differentially expressed between the three sample groups (Supplementary Table S2). The protein expression changes were considered significant when p value < 0.05 and when more than a 2-fold difference was detected between the compared sample groups. Principal component and hierarchical cluster analyses of the expression profiles of the 91 differentially expressed proteins resulted in three distinct clusters, as shown in Figure 4A. The representations of trends in the expression changes of some of these proteins between MERS-CoV patients and control subjects are illustrated in Figure 4B.
 |
Figure 4 (A) PCA and hierarchical cluster analysis using the expression dataset of 91 differentially expressed proteins between MERS-CoV patients and control subjects. The expression changes of these proteins separate the samples into three distinct clusters, indicating the potential use of these proteins for accurate stratification and as prognostic biomarkers for MERS-CoV patients (see Supplementary Table S2). (B) The representations of the expression changes of some of these proteins between MERS-CoV patients and control subjects. The normalized quantitative values of individual proteins across all sample groups are depicted as box plots using the Qlucore Omics Explorer version 3.7 (Lund, Sweden) (https://qlucore.com). |
Identification of Biomarkers Implicated in Pathogenesis and Regulation of Various Pathways in the Two Sub-Types of Coronaviruses, SARS-CoV-2 and MERS-CoV
We analyzed two extreme groups of MERS-CoV patients, namely, those with severe disease who were admitted to the ICU and died of the disease and those who recovered from the infection. In addition, we used healthy control subjects without a history of MERS-CoV for baseline comparisons.
We evaluated the intersection between 193 and 91 differentially expressed proteins between COVID-19 and MERS-CoV cohorts, respectively. Only 49 proteins overlap between the two datasets, indicating some level of homogeneity in the protein expression profiles associated with the two implicated coronaviruses. The expression profiles of the 49 proteins could distinguish the sub-groups of the two COVID-19 and MERS-CoV sample cohorts (Figure 5A). The list and cellular processes of some of these proteins are as listed in Supplementary Tables S3 and S4. In addition, changes in the abundance of some of the proteins are illustrated in Figure 5B.
 |
Figure 5 (A) Identification of biomarkers implicated in regulation of SARS-CoV-2 and MERS-CoV. (Top panel) The Venn diagram showing 49 proteins that overlap between the two datasets of COVID-19 and MERS-CoV, indicating some level of homogeneity in protein expression profiles of the two implicated coronaviruses. PCA plot (left lower panel) and hierarchical cluster analysis (right lower panel) using the expression dataset of the 49 proteins that overlap between the two datasets of COVID-19 and MERS-CoV. (B) Representative changes in abundance of some of the 49 proteins between the different grades of COVID-19 patients. The normalized quantitative values of individual proteins across all the sample groups are depicted as box plots, which were generated using the using the Qlucore Omics Explorer version 3.7 (Lund, Sweden) (https://qlucore.com). |
Discovery/Development of COVID-19 Biomarker Protein Panel for Objective Prediction of Disease Prognosis
We compared the most significant canonical pathways between the COVID-19 and MERS-CoV datasets using comparative IPA. This analysis enables the mapping of molecules from the two datasets and displays significant associations with canonical pathways.
It further allows for assessments or predictions of the roles of the implicated molecules to determine whether they act as either activators or inhibitors of canonical pathways, irrespective of their expression status. The hierarchical clustering heat maps show that both COVD-19 and MERS-CoV are involved in similar canonical pathways, but with greater enrichment among COVID-19 samples than MERS-CoV. We also observed that certain organs are more significantly perturbed in COVID-19 than MERS samples, including the liver and kidney (Supplementary Figure 3A–C).
Discovery/Development of Potential Protein Panel for Diagnosis of Asymptomatic Cases and Prognosis of COVID-19 Disease Progression
We further evaluated the expression signatures of the 49 above-mentioned differentially expressed proteins to assess their potential use for objectively predicting the progression of COVID-19 patients between asymptomatic, combined non-severe disease, and severe illness. The majority of the 49 differentially expressed peripheral blood proteins were upregulated in severely ill patients compared with non-severe cases. Only 9 of the 49 proteins were upregulated in asymptomatic COVID-19 samples compared with the other groups. On the other hand, 12 of the 49 proteins were highly expressed in patients who recovered from severe cases of COVID-19 compared with severely ill patients with a fatal outcome and patients with non-severe disease. The remaining 28 of the 49 proteins were highly expressed in patients who died from severe cases of COVID-19 compared with the other samples. These proteins, therefore, could be potential COVID-19 diagnostic markers for asymptomatic cases and prognostic markers for severely ill patients who will recover and those with a fatal outcome of the disease. The gene names and fold changes in expression levels between patients with non-severe and severe illness are indicated in Figure 6A, and a heat map of the expression profiles of these potential COVID-19 biomarkers is presented in Figure 6B.
The list and functional annotations/disease associations of some of the proteins involved in coronavirus infections and other cellular processes are described in Supplementary Table S3, S4 and Supplementary Figure 3A–C.
Discussion
The ability to identify biological markers, including key protein–protein interactions, involved in the response to different coronavirus infections may improve disease management. Several studies to date have shown that, although people from all age groups are vulnerable to infection by SARS-CoV-2, certain COVID-19 patients are at higher risk, particularly the older population and patients with comorbidities.5,25 Previous studies have also shown that the disease course after MERS-CoV exposure is unique26 and distinct from that of SARS-CoV-2. Moreover, the re-emergence of coronavirus outbreaks indicates their capacity to mutate or re-combine to become infectious and pathogenic across species barriers and may potentially result in future pandemics. Since genetic changes in viruses are part of their evolutionary process, the host responses to these viruses may vary at the molecular pathway level, including protein expression. Therefore, the aim of this study was to explore the detailed molecular mechanisms of the host immune response to SARS-CoV-2 among groups with different disease severity and further compare host responses between SARS-CoV-2 and MERS-CoV. To our knowledge, a comparative analysis of the molecular pathways of host responses to SARS-CoV-2 and MERS-CoV has not been reported before. The findings of this study could be critical for precision medicine and the discovery of new therapeutic targets, as well as for the development of significant cost-effective and life-saving applications.
Here, we show that patients infected with SARS-CoV-2 display a molecular signature that corresponds to the severity of the disease and involves a number of biological pathways. Liver X receptor (LXR) activation, acute-phase reaction (APR), and production of nitric oxide and reactive O2 species (NO-ROS) are markedly affected in samples from severe COVID-19 cases and are able to differentiate severely ill patients who recovered from the disease from patients with a fatal outcome of the disease, while asymptomatic, mild, and moderate cases showed unique expression profiles that differed from those of severely ill patients. The liver X receptors (LXRs) and agonists (RXRs) play central roles in the transcriptional control of lipid metabolism. They have also been found to modulate immune and inflammatory responses in macrophages. The ability of LXRs to integrate metabolic and inflammatory signals makes them particularly attractive targets for intervention not only in metabolic diseases but also in response to viral infection.27,28 APR is a prominent systemic reaction of the organism to local or systemic disturbances in its homeostasis caused by infection, tissue injury, trauma or surgery, neoplastic growth, or immunological disorders, and it involves responses from a large number of pro-inflammatory cytokines.28,29 In response to viral infection, this pathway may have a role in mediating the severity of symptoms or causing the cytokine storm response (Figure 3A). Interestingly, two recent genome-wide studies indicate that changes in the genetic makeup of certain cytokines are associated with disease severity.30 NO-ROS bioavailability and increased expression are implicated in damage to cardiovascular tissues.31 The occurrence of severe SARS-CoV-2 infection and a high risk of poor outcomes of COVID-19, particularly in patients with comorbidities (including hypertension and diabetes), combined with the observation of NO-ROS pathway involvement in severely ill (recovered and deceased) patients, highlights the importance of this pathway as a COVID-19 severity marker.
Several studies have applied proteomic approaches to discover blood biomarkers for COVID-19.12–14 However, differences in the analytical platforms, pre-analysis sample handling, cohort of patients, and the timing of sample collection are among the factors that limit inter-laboratory comparisons of results. In Park et al,13 the authors compared their COVID-19 proteomic signature with those described by Messner et al and Shen et al,12,14 and they reported that approximately 71% of identified proteins overlapped among the three different datasets.13 In spite of this relative homogeneity, only seven (7) differentially expressed proteins were common between Part et al and Shen et al, with varying expression changes between the analyzed sample cohorts.13 However, the third study (Messner et al) described 27 differentially expressed proteins between COVID-19 patients requiring oxygen support and critical cases not requiring oxygen. Despite the above-listed confounding factors in inter-laboratory comparisons of proteomic data, 22 of 27 previously reported proteins were among the 49 potential COVID-19 biomarkers identified in this study.
Only 9 of the 49 proteins were upregulated in asymptomatic samples compared with the other groups. On the other hand, 12 of the 49 proteins were highly expressed in patients who recovered from a severe case of COVID-19 compared with those with a fatal outcome and other groups, and the remaining 28 of the 49 proteins were highly expressed in patients who died of severe COVID-19 compared with the other samples. These proteins, therefore, could be potential COVID-19 diagnostic markers for asymptomatic cases and prognostic markers for severely ill patients who will recover and those with a fatal outcome of the disease. Unlike previously reported proteomic data that were focused on disease severity, our observed biomarkers have potential applications for the diagnosis of asymptomatic cases (9 markers) as well as prognosis of severely ill patients, with 12 and 28 markers for patients who recovered and those with a fatal outcome of the disease, respectively. Furthermore, the additional uniqueness of this study is the utility of these markers in MERS-CoV patients in addition to SARS-CoV infections.
This study also shows that the host response to SARS-CoV-2 is markedly different from the host response to MERS-CoV. The results of the clustering analysis of MERS-CoV vs SARS-CoV-2 indicate a MERS-CoV-specific profile that differs from the SARS-CoV-2 profile, affecting a number of canonical/essential pathways as well as organ-associated pathways. The host responses to the two types of infections involve 49 differentially expressed proteins, with unique expression profiles that differentiate the two infections. NO-ROS, LXRs, and APR protein expression levels generate a SARS-CoV-2 cluster that is distinct from MERS-CoV. Similarly, the expression levels affecting vital organs such as the liver and kidney (Supplementary Figure 2) indicate that the response to MERS-CoV across different organ-dependent pathways differs from the response to SARS-COV-2. There was a differential expression profile distinct to MERS-CoV patients in a number of pathways involving the injury of tissues, particularly the kidney and liver, in which MERS-CoV expression was lower than that of SARS-CoV-2.
We also investigated whether SARS-CoV-2 virus particles are circulating in the blood and whether this is associated with disease symptoms, severity, or outcome. Coronaviruses are not blood-borne diseases; rather, they are usually transmitted by direct contact with infectious pathogens via the upper or lower respiratory tract. Despite this transmission mode, viral shedding in bodily fluids such as plasma or serum has been detected.18–21 We confirmed positive viral shedding in 6 (11.76%) of the 51 peripheral blood samples analyzed. Therefore, medical and laboratory staff need to strictly adhere to safety protocols when handling coronavirus samples and particularly COVID-19 patients. Furthermore, such safety precautions should be implemented to avoid the risk of transmitting coronaviruses through the transfusion of infected blood products.18,19
Elevated concentrations of immunological markers (eg, interleukins) are consistent with the “cytokine storm” hypothesis.32 A number of studies have reported associations between protein concentrations and disease severity, and predictive algorithms have begun to be formulated on this basis. The first mass-spectrometry-based proteomic analyses of patients’ blood were reported,33,34 and the picture emerging from these studies is one of systemic perturbation. There is now a need for clinical sensitivity and specificity for any specific biomarker signature associated with COVID-19 used to make clinical decisions. Thus, this initial assessment of major protein components in samples from COVID-19 patients and comparison with those from MERS-CoV shed light on SARS-CoV-2-specific host response pathways. Based on these findings, we propose the rapid deployment of plasma proteomics or throat swab proteomics in a well-regulated discovery proteomics environment with sufficient samples to lend power to studies; this will be a key part of future developments in combatting this disease. The overall objective can include genomic data and should certainly include advanced health informatics approaches to translate data into clinically useful information.
Conclusion
Multiple protein panels have been described as promising COVID-19 biomarkers. Surprisingly, only a handful of them overlap among the different published studies, and none of them have been validated as protein panels for routine clinical diagnostics. In the present study, the peripheral blood of patients with different severity grades of SARS-CoV-2 and MERS-CoV infections were analyzed to determine whether there were distinct molecular changes characteristic of disease outcomes. The benefit and uniqueness of our study lie in the discovery of objective markers that may be used to predict COVID-19 outcomes at the point of care and protein panels to identify asymptomatic individuals, those who will progress to develop mild, moderate, or severe disease, and those who will recover without life-threatening complications of COVID-19 disease. The results reveal detailed molecular mechanisms of the host immune response to SARS-CoV-2 in different diagnosed groups and further indicate how the SARS-CoV-2 host response compares to the MERS-CoV host response. Taken together, the findings in this study support some of the previously described potential COVID-19 protein biomarkers and further define new additional diagnostic markers for the identification of asymptomatic individuals. The results also offer a means of objectively predicting disease outcomes among severely ill COVID-19 patients. Finally, we detected viral shedding in more than 11% of analyzed peripheral blood samples of these patients, highlighting the need for caution to avoid the possible risk of transfusion of infected blood products.
Abbreviations
MERS-CoV, Middle East respiratory syndrome coronavirus; SARS-CoV-2, Severe Acute respiratory syndrome coronavirus-2; COVID-19, Disease caused by SARS-CoV-2.
Data Sharing Statement
Details of the materials, methodologies, and all data produced, analyzed, and reported in this study are incorporated in this manuscript and the additional corresponding Supplementary Data Files. The accession number and gene name used for species-specific protein identification and description reported in this study are in the UniProt/FASTA format. The lab logs on sample preparation and raw mass spectrometry data files produced by Waters Synapt G2 HDMSE will be made available by the lead contact/corresponding author upon reasonable request.
Ethics Approval
The study was approved by the institutional research review committee of King Faisal Specialist Hospital and Research Centre (RAC#2200011).
Consent to Participate
The study was approved by the institutional research review committee of King Faisal Specialist Hospital and Research Centre. All patients provided informed consent, and the study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable international ethical standards.
Acknowledgments
We acknowledge King Faisal Specialist Hospital and Research Centre (General Organization), Riyadh KSA, for funding, resources, and support. The support of the administration at King Faisal Specialist Hospital and Research Centre is highly appreciated. We would like to thank the Infectious Disease staff and physicians for their help in patient selection and recruitment for this study. We thank Melvin Velsaco Zabalerio for technical assistance with graphic designs.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
King Faisal Specialist Hospital and Research Centre (Saudi Arabia).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Zumla AI, Memish ZA. Middle east respiratory syndrome coronavirus: epidemic potential or a storm in a teacup? Eur Respir J. 2014;43(5):1243–1248. doi:10.1183/09031936.00227213
2. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi:10.1056/NEJMoa1211721
3. Azzi L, Baj A, Alberio T, et al. Rapid salivary test suitable for a mass screening program to detect SARS-CoV-2: a diagnostic accuracy study. J Infect. 2020;81(3):e75–e78. doi:10.1016/j.jinf.2020.06.042
4. Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45–e50. doi:10.1016/j.jinf.2020.04.005
5. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi:10.1016/S0140-6736(20)30183-5
6. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi:10.1016/S0140-6736(20)30251-8
7. Vellingiri B, Jayaramayya K, Iyer M, et al. COVID-19: a promising cure for the global panic. Sci Total Environ. 2020;725:138277. doi:10.1016/j.scitotenv.2020.138277
8. Devaux CA, Rolain JM, Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53(3):425–435. doi:10.1016/j.jmii.2020.04.015
9. Li K, Wu J, Wu F, et al. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Invest Radiol. 2020;55(6):327–331. doi:10.1097/RLI.0000000000000672
10. Sun Y, Koh V, Marimuthu K, et al. Epidemiological and clinical predictors of COVID-19. Clin Infect Dis. 2020;71(15):786–792. doi:10.1093/cid/ciaa322
11. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi:10.1016/S0140-6736(20)30566-3
12. Messner CB, Demichev V, Wendisch D, et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11(1):11–24 e14. doi:10.1016/j.cels.2020.05.012
13. Park J, Kim H, Kim SY, et al. In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients. Sci Rep. 2020;10(1):22418. doi:10.1038/s41598-020-80120-8
14. Shen B, Yi X, Sun Y, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72 e15. doi:10.1016/j.cell.2020.05.032
15. Lindoso RS, Sandim V, Collino F, et al. Proteomics of cell-cell interactions in health and disease. Proteomics. 2016;16(2):328–344. doi:10.1002/pmic.201500341
16. Alaiya AA, Aljurf M, Shinwari Z, et al. Protein signatures as potential surrogate biomarkers for stratification and prediction of treatment response in chronic myeloid leukemia patients. Int J Oncol. 2016;49(3):913–933. doi:10.3892/ijo.2016.3618
17. Molinari N, Roche S, Peoc’h K, et al. Sample pooling and inflammation linked to the false selection of biomarkers for neurodegenerative diseases in top-down proteomics: a pilot study. Front Mol Neurosci. 2018;11:477. doi:10.3389/fnmol.2018.00477
18. Chang L, Yan Y, Wang L. Coronavirus disease 2019: coronaviruses and blood safety. Transfus Med Rev. 2020;34(2):75–80. doi:10.1016/j.tmrv.2020.02.003
19. Kampf G, Todt D, Pfaender S, Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect. 2020;104(3):246–251. doi:10.1016/j.jhin.2020.01.022
20. Leclercq I, Batejat C, Burguiere AM, Manuguerra JC. Heat inactivation of the middle east respiratory syndrome coronavirus. Influenza Other Respir Viruses. 2014;8(5):585–586. doi:10.1111/irv.12261
21. Pyankov OV, Bodnev SA, Pyankova OG, Agranovski IE. Survival of aerosolized coronavirus in the ambient air. J Aerosol Sci. 2018;115:158–163. doi:10.1016/j.jaerosci.2017.09.009
22. Alkhayal Z, Shinwari Z, Gaafar A, Alaiya A. Proteomic profiling of the first human dental pulp mesenchymal stem/stromal cells from carbonic anhydrase II deficiency osteopetrosis patients. Int J Mol Sci. 2020;22(1):380. doi:10.3390/ijms22010380
23. Colak D, Alaiya AA, Kaya N, et al. Integrated left ventricular global transcriptome and proteome profiling in human end-stage dilated cardiomyopathy. PLoS One. 2016;11(10):e0162669. doi:10.1371/journal.pone.0162669
24. Al-Saud H, Al-Romaih K, Bakheet R, et al. Automated SARS-COV-2 RNA extraction from patient nasopharyngeal samples using a modified DNA extraction kit for high throughput testing. Ann Saudi Med. 2020;40(5):373–381. doi:10.5144/0256-4947.2020.373
25. Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi:10.1136/bmj.m1091
26. Mackay IM, Arden KE. MERS coronavirus: diagnostics, epidemiology and transmission. Virol J. 2015;12:222. doi:10.1186/s12985-015-0439-5
27. Zelcer N, Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116(3):607–614. doi:10.1172/JCI27883
28. Gruys E, Toussaint MJ, Niewold TA, Koopmans SJ. Acute phase reaction and acute phase proteins. J Zhejiang Univ Sci B. 2005;6(11):1045–1056. doi:10.1631/jzus.2005.B1045
29. Gruys E, Toussaint MJ, Upragarin N, et al. Acute phase reactants, challenge in the near future of animal production and veterinary medicine. J Zhejiang Univ Sci B. 2005;6(10):941–947. doi:10.1631/jzus.2005.B0941
30. Zhang Q, Bastard P, Liu Z, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:6515. doi:10.1126/science.abd4570
31. Matsubara K, Higaki T, Matsubara Y, Nawa A. Nitric oxide and reactive oxygen species in the pathogenesis of preeclampsia. Int J Mol Sci. 2015;16(3):4600–4614. doi:10.3390/ijms16034600
32. Soy M, Keser G, Atagunduz P, Tabak F, Atagunduz I, Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. 2020;39(7):2085–2094. doi:10.1007/s10067-020-05190-5
33. Bojkova D, Klann K, Koch B, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583(7816):469–472. doi:10.1038/s41586-020-2332-7
34. Shu T, Ning W, Wu D, et al. Plasma proteomics identify biomarkers and pathogenesis of COVID-19. Immunity. 2020;53(5):1108–1122 e1105. doi:10.1016/j.immuni.2020.10.008
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.