Back to Journals » Cancer Management and Research » Volume 12
Allicin Inhibits Proliferation by Decreasing IL-6 and IFN-β in HCMV-Infected Glioma Cells
Authors Yang Z, Du J, Zhu J, Rong Y, Chen S, Yu L, Deng X, Zhang X, Sheng H, Yang L , Lu X, Li D, Yin B , Lin J
Received 3 May 2020
Accepted for publication 10 July 2020
Published 17 August 2020 Volume 2020:12 Pages 7305—7317
DOI https://doi.org/10.2147/CMAR.S259677
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Zelin Yang,1,* Jizao Du,2,* Jinjin Zhu,3 Yuxi Rong,1 Shaohuai Chen,1 Lisheng Yu,1 Xiangyang Deng,1 Xiaojia Zhang,1 Hansong Sheng,1 Liang Yang,1 Xiangqi Lu,1 Dandong Li,1 Bo Yin,1 Jian Lin1
1Department of Neurosurgery, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, Wenzhou, People’s Republic of China; 2Digestive Cancer Center, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, People’s Republic of China; 3Department of Neonatology, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, Wenzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jian Lin
The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, 109 Xueyuan Road, Wenzhou, Zhejiang, People’s Republic of China
Tel +86 577 8800 2502
Fax +86 577 8883 2693
Email [email protected]
Purpose: Allicin, an extract of garlic, has antitumor effects in multiple tumor types. However, the efficacy of allicin for treating glioblastoma has not yet been examined. This study examined the antitumor effect of allicin on human cytomegalovirus (HCMV)-infected glioblastoma multiforme (GBM) and its role in cytokine signaling.
Materials and Methods: HCMV-infected glioblastoma was modeled by transfection of U87MG glioblastoma cells with HMCV proteins. MTT assay was used to assess the effect of allicin on the proliferation of glioma cells. Western blot analysis was used to detect the effect of allicin on the expression of intermediate-early gene 2 (IE2) and p53. Reverse transcription-quantitative polymerase chain reaction was used to assess and the levels of interleukin (IL)-6 and interferon (IFN)-β. Single cell gel electrophoresis was used to analyze changes in radiotherapy-induced DNA damage.
Results: Transfection of the IE2 protein led to decreased p53 expression and increased glioblastoma cell proliferation. Allicin inhibited this proliferation in a dose- and time-dependent manner. An inhibitory effect on cytokine release was observed in GBM cells treated with allicin. After treatment with allicin, p53 levels increased significantly, whereas expression of the inflammatory factors such as IL-6 and IFN-β decreased. U87MG cells treated with allicin and 10 Gy irradiation had increased intracellular DNA damage compared to either treatment alone.
Conclusion: Allicin inhibited proliferation of glioblastoma cells in vitro. Allicin also inhibited cytokine release, upregulated p53 activity, and increased the sensitivity of glioblastoma to radiotherapy. These results suggest that allicin is effective against HCMV-infected glioblastomas.
Keywords: allicin, cytomegalovirus, glioblastoma, inflammatory mediators, proliferation, radiotherapy
Introduction
Subdermal glioblastoma multiforme (GBM) is one of the most malignant and invasive tumors in the adult central nervous system and accounts for approximately 50% of all intracranial tumors.1–3 Although current surgical resection methods are feasible with a combination of radiotherapy or chemotherapy, the median survival is only about 1 year.4 Viral genes and proteins are detected in a high percentage of both high-grade and low-grade gliomas.5 Further, multiple reports confirm that human cytomegalovirus (HCMV) gene and protein expression is detected in >90% of GBM cells but not adjacent normal brain or other neuropathologies, suggesting a potential link between glioma and HCMV infection.6,7 Brain as the preferred site of HCMV infection, which can lead to abnormal differentiation of neural stem cells or inhibit the differentiation to normal astrocytes.8,9 Meanwhile, HCMV infection also induces glioma stem cell (GSCs) phenotype, which expresses the same marker CD133 as neural stem cells. HCMV infection leads to increased expression of GSCS markers (Sox2, Notch1, Nestin, Oct4) such as CD133, and inhibits the differentiation of GCSCs.10 HCMV infection is persistent worldwide; it causes lifelong infection in 40–90% of adults.11 Increasing reports about the specific expression of HCMV in human malignant tumors suggest the possibility that HCMV may be beneficial to tumor progression.12 Immediate-early (IE) genes are the first viral genes activated after HCMV infection. IE86 is a strong transactivator produced by viral and cellular gene expression that interacts with cellular transcription factors.17 It can be involved in viral and cellular promoter repression through positive or negative regulation. Furthermore, the expression of IE86 is also related to the expression of activating transcription factor 5(ATF5), P300, and acetylated histones, indicating that the overexpression of the HCMV IE86 protein is correlated with malignancy and histone acetylation in glioma.13,14
Interferons (IFNs) are cytokines induced after viral infection.23 They can induce the expression of mucovirus protein A, a cytoplasmic GTPase with antiviral activity, which plays a vital role in protecting cells from viral infection.24 Interleukin (IL)-6 is a regulator of the host innate and humoral immune responses. IL-6 is also induced during viral infection, including HCMV infection.25–27 Interestingly, IFN-β and IL-6 are expressed in glioblastomas and play an important role in tumor proliferation and migration.23,28
The presence of stromal cells in the tumor microenvironment is one of the main reasons for the high recurrence rate in glioma. Macrophages, one of the stromal cells in glioma, can release the chemokines CCL5 and CCL2 to guide monocytes, macrophages, and mesenchymal stem cells to infiltrate into tumors and participate in tumor proliferation and migration. In addition, when IL-6, IFN-β, and other inflammatory factors are highly expressed in macrophages, the transformation from M1-type to M2-type can be accelerated, which exacerbates the infiltration of macrophages into the tumor.
Allicin (2-propene-1-sulfinothioic acid S-2-propenyl ester) is a compound extracted from freshly crushed garlic. Due to its unique antibacterial and anti-inflammatory effects, it may have pro-apoptotic ability, suggesting its use as a cancer therapy.15–17 The anticancer effects of allicin have been examined in vitro in lung cancer, hepatocellular carcinoma, gastric cancer, and breast cancer cell lines.18–20 Although it is known that allicin-induced glioma cell apoptosis is regulated by MAPK/ERK-dependent pathways,21,22 the efficacy of allicin for treating glioblastoma has not yet been examined.
We hypothesized that overexpression of IFN-β and IL-6 in glioblastoma is due to HCMV infection and that allicin could treat glioblastoma by reducing inflammatory factors released by its unique anti-inflammatory and antiviral mechanisms. Therefore, the purpose of this study was to elucidate the effect of allicin on the proliferation of U87MG glioblastoma cells, specifically examining inflammatory cytokines and macrophages. We also examined whether allicin can inhibit proliferation and increase the sensitivity of glioblastoma cells to radiotherapy in vitro.
Materials and Methods
Immunohistochemistry
Paraffin-embedded glioblastoma sections were provided by the Department of Pathology at the Second Affiliated Hospital of Wenzhou Medical University. The paraffin sections were heated at 60°C for 2 hours and then incubated in 100% xylenes for 5 minutes for dewaxing; this process was repeated twice. Slides were then rehydrated in an ethanol gradient (100%, 75%, 50%, and 0% ethanol) for 5 minutes. The specimens were then placed in 100°C sodium citrate buffer (pH = 6) and microwaved for 45 minutes for antigen repair. The endogenous peroxidase activity was blocked with 1% hydrogen peroxide solution, and slides were washed three times in phosphate-buffered saline (PBS) (pH = 7.2). The slides were incubated at 37°C for 1 hour, at which point they were incubated with primary antibody (anti-IE2, Millipore, 1:100). Goat serum was applied for 2 hours. A mouse two-step test kit (PV-9002, ZSGB-BIO) was combined with the secondary antibody for 30 min, and then DAB (Solarbio, China) was added for 10 min. The slides were then washed with water to stop the reaction, dehydrated in an alcohol gradient, and placed in xylenes to increase transparency. This study obtained the written informed consent of the patients.
Cell Culture
U87MG and THP1 (Bnbio, Beijing, China) cells were grown in DMEM medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Gibco). Cells were seeded in Petri dishes in a 5% humidified incubator at 37°C. The cells were cultured for 2–3 days until confluence, trypsinized, and seeded in 6-well or 96-well tissue culture plates. The serum was removed from the culture medium for 1–2 days before adding various reagents.
Transfection of pEGFP/IE2 Expression Plasmid
U87MG and THP1 cells were seeded in 12-well culture plates and cultured to 90% confluence with fresh medium. The pEGFP/IE2 plasmid was then transfected into U87MG and THP1 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The treated cells were compared with cells transfected with pEGFP/IE2. The expression of IE2 was monitored by Western blot analysis 48 hours after transfection.
Western Blot Analysis
Cells were lysed on ice with RIPA buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 2 mM PMSF, 0.05 mM Gasstatin A, 0.2 mM Leupeptin) and washed with PBS. The lysate was centrifuged at 12,000 rpm for 10 minutes at 4°C. Fifty micrograms of protein was boiled in reduced SDS loading buffer, and the samples were loaded on a 10% SDS-polyacrylamide gel for electrophoresis and transferred onto an Immobilon PVDF membrane (Millipore). After blocking with 5% skim milk for 2 hours at 37°C, the membrane was incubated with the antibody against IE (Millipore) at 4°C overnight, washed, and then incubated with a horseradish peroxidase-conjugated secondary antibody. ECL was used to detect immunoreactive protein.
MTT Assay
U87MG cells were seeded in 96-well plates at 500 cells per well and treated with allicin. After allicin treatment, cells were washed with PBS and incubated with DMEM containing 0.5 mg/mL MTT [3- (4,5-dimethylthiazole) −2-yl] −2,5-diphenyltetrazole bromide] for 2 hours at 37°C. The medium was removed, and DMSO was added. Cell viability was evaluated by measuring absorbance at 570 nm with 680 microplate reader (microplate reader) (Germany).
Reverse Transcription-Quantitative Polymerase Chain Reaction
RNA from U87MG cells treated with allicin was isolated using TRIzol reagent (Solarbio, China). The Bio-Rad real-time PCR system (Hercules, CA, USA) was used for PCR. Thermal cycling conditions were as follows: 95°C for 30 seconds, then 40 cycles at 95°C for 5 seconds, 60°C for 30 seconds, and 72°C for 15 seconds. The following primers were used for PCR: IL-6 (F: 5ʹ-ACAACCACGGCCTTCCCTACTT-3ʹ, R: 5ʹ-CACGATTTCCCAGAGAACATGTG-3ʹ), IFN-β (F: 5ʹ-ACGCCGCATTGACCATCTAT-3ʹ, R: 5ʹ-GTCTCATTCCAGCCAGTGCT-3ʹ), IE2 (F: 5′-GCGCAATATCATGAAAGATAAGAACA-3′, R: 5′-GATTGGTGTTGCGGAACATG-3′), TP53 (F: 5′-CGCCTGAGGTTGGCTCTGACTGTA-3′, R: 5′-GTCTCTCCCAGGACAGGCACAAAC-3′), and GAPDH (F: 5ʹ-GCTCCTCCT-GTTCGACAGTCA-3ʹ, R: 5ʹ-ACCTTCCCCATGGTGTCTGA-3ʹ). The SYBR PrimeScript RT-PCR kit (Takara, Bio, Inc., Otsu, Japan) was used to assay the IL-6 and TNF-β mRNA expression levels. Relative expression levels were analyzed using the 2−ΔΔCq method.
ELISA
U87MG cells were seeded on 12-well plates 12 hours before the experiment and treated with or without allicin at a concentration of 60 mg/mL. After 24 hours, the supernatant was collected and centrifuged at 10,000 rpm and 4°C for 10 minutes. The IFN-β and IL-6 concentrations were measured using a quantitative ELISA kit according to the manufacturer’s instructions (MultiSciences, Hangzhou, China).
Single Cell Gel Electrophoresis (Comet) Assay
Cells were grown in an incubator for 6 hours, then irradiated with 10 Gy irradiation (dose rate: 1.1 Gy/min) using a CHF 320 G generator (Gilardoni, Mandello del Lario, Italy). After irradiation, the cells were thoroughly washed three times with sterile PBS and incubated with a 0.25% trypsin solution for 4 minutes at 37°C. After termination of digestion, cells were centrifuged at 200 g, then 20 μL of the cells was combined with 180 μL of 0.7% low-melting point agarose in PBS without Ca or Mg and immediately transferred to ground glass slides precoated with 1% normal-melting point agarose in PBS. The coverslips were stored at 4°C to solidify the agarose. Then, the coverslips were placed in an alkaline lysis buffer (1% Triton X-100 and 10% dimethyl sulfoxide) for 45 minutes at 4°C in the dark. After lysis, the slides were washed with running buffer (1 mM Na2EDTA and 300 mM NaOH, pH = 13) for 10 minutes and then placed in a horizontal electrophoresis device containing the same running buffer for 20 minutes. DNA was fixed in 4°C PBS for 15 min. Subsequently, the slides were gently washed in a neutralization buffer solution for 5 minutes (0.4 M Tris-HCl, pH = 7.5), dehydrated through an ethanol series (70%, 85%, and 100%), dried at room temperature, and stored. For microscopic analysis, slides were stained with 2 mg/mL ethidium bromide in distilled water. Unless otherwise stated, all chemicals were purchased from Solarbio (Beijing, China).
Statistical Analysis
Experimental data are expressed as mean ± standard error from three replicates. The statistical difference between allicin treatment and vehicle treatment was compared by paired T test. Statistical software SPSS 21.0 (SPSS, Inc. USA) was used to analyze statistical differences between groups using ANOVA. GraphPad Prism (Version 7.0; GraphPad Software, Inc. USA) was used to produce the graphics. Statistical significance was defined as P < 0.05.
Results
IE2 Promotes Proliferation and Inflammatory Cytokine Expression in U87MG Cells
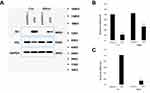
Immunohistochemistry was used to confirm the presence of HCMV proteins in patient glioma samples (Figure 1A). IE2 was expressed in paraffin-embedded sections from glioma patients but not in peritumoral tissues. We then used the U87MG glioma cell line to determine whether HCMV infection affects the expression level of inflammatory cytokines in glioma cells. There was decreased p53 and IE2 expression (Figure 1B) and increased IL-6 and IFN-β mRNA expression in HCMV-infected glioma cells (Figure 1D and E). MTT assay showed that IE2 transfection could promote the proliferation of glioma cells (Figure 1C). Therefore, our results demonstrate that HCMV is present in glioma tissues and IE2 expression can increase the malignancy of gliomas by affecting the expression of p53, IL-6, and IFN-β.
High Concentrations of Allicin Reduce IE2 Protein Expression
We treated U87MG cells with a concentration gradient of allicin (10 μg/mL, 30 μg/mL, and 60 μg/mL) to determine the optimal concentration. Although the lower concentrations (10 μg/mL and 30 μg/mL) of allicin were able to decrease IE2 protein expression in U87MG cells, the higher allicin concentration (60 μg/mL) caused the largest decrease in IE2 levels (Figure 2A). These results were also validated at the mRNA level (Figure 2B). Therefore, all subsequent experiments used 60 μg/mL allicin.
Allicin Inhibits the Proliferation of U87MG Cells
MTT assay was used to assess the impact of allicin on the proliferation of U87MG cells. When treated with a specific allicin concentration (60 μg/mL), U87MG cell proliferation was decreased in a time-dependent manner (Figure 3).
Allicin Reduces IE2 and P53 Protein Expression in U87MG Cells
According to a previous study, IE2 can directly bind to p53 and increase its expression.29 Therefore, we performed Western blotting to assess p53 and IE2 protein expression in cells treated with allicin. After treatment with 60 μg/mL allicin, the p53 levels increased significantly and the IE2 levels decreased (Figure 4A). Further analysis by RT-qPCR showed that the changes in TP53 and IE2 mRNA were statistically significant (Figure 4B and C). Combined with the previous MTT assay, these results indicate that allicin may increase p53 expression by reducing IE2 protein levels, thereby inhibiting the proliferation of glioma cells.
Allicin Reduces Inflammatory Responses in HCMV-Infected Glioma Cells
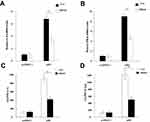
RT-qPCR results showed that allicin regulates inflammatory cytokines in glioma cells. Allicin did not significantly decrease IL-6 or IFN-β in U87MG cells that did not express IE2. In contrast, in glioma cells with overexpression of IE2, the expression of IL-6 and IFN-β was increased, and allicin had a significant inhibitory effect on the expression of these two inflammatory factors (Figure 5A and B). In addition, we found that the release of the proinflammatory cytokines IFN- and IL-6 decreased after treatment with allicin (Figure 5C and D). These data show that allicin is effective in preventing the release of inflammatory cytokines.
Allicin Increases the Sensitivity of Glioma Cells to Radiotherapy
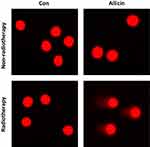
Comet assay was used to assess DNA damage of U87MG cells after radiotherapy. Little DNA damage was observed in untreated U87MG cells or cells treated with allicin or radiotherapy alone. The results suggested that glioma cells were resistant to radiotherapy. However, U87MG cells treated with allicin and 10 Gy irradiation had increased intracellular DNA damage (Figure 6).
Effect of Allicin on Macrophage Differentiation
We transfected IE2 plasmid into macrophages and treated them with allicin and observed results similar to those in glioma cells (Figure 7). IE2 promoted IL-6 and IFN-β mRNA and inhibited p53 expression in macrophages, as well as enhanced proliferation (Figure 8A and B). After treatment with allicin (60 μg/mL), IL-6 and IFN-β mRNA expression in macrophages decreased (Figure 8C and D), whereas the p53 protein and mRNA levels increased (Figure 7D and E). IL-6 and IFN-β secretion were also reduced (Fig. E and F). Further, the proliferation of macrophages was inhibited, and the sensitivity to radiotherapy was enhanced with allicin treatment (Figure 9). This suggests that allicin reduces the level of intracellular inflammatory factors into macrophages.
Discussion
Overall, our results showed that allicin could effectively inhibit the proliferation of U87MG glioma cells. Allicin inhibited the expression of the inflammatory cytokines IL-6 and IFN-β in glioma cells and macrophages and increased the expression of p53 at the protein level, thereby inhibiting the proliferation of glioma cells. Future studies should evaluate allicin as a new antitumor agent in the treatment of glioma.
Glioblastoma is one of the most common invasive brain cancers. Most patients have a poor prognosis, with an average 1-year survival time.30 Although many new medicines for glioblastoma have been discovered, most current treatments are ineffective and have serious side effects.31 There is increasing evidence that HCMV plays an important role in promoting the formation of GBM and its malignant biological behaviors by increasing the expression of inflammatory factors in tumor cells.32,33 Therefore, it has become a major research direction to identify anticancer and antiviral drugs from gene therapy, biological therapy, and natural foods for the treatment of GBM.34 Allicin, a sulfur-derived compound in garlic, has antiviral and anticancer effects. However, the mechanism of its action remains unclear.35,36 Previous studies have shown that allicin has therapeutic effects on influenza virus and herpes virus.37,38 In addition, allicin can reduce the risk of the development of cancers such as melanoma and leukemia, inhibit the growth of a variety of cancer cells, and may be an important anticancer agent.20,39,40 Therefore, allicin has the potential to inhibit the proliferation of GBM.
The main findings of this study are the optimal dose of allicin in reducing IE2 expression in U87MG glioma cells infected with HCMV and the inhibitory effect of allicin on these cells. The effect of allicin on intracellular inflammatory cytokines in U87MG cells infected with HCMV was dose-dependent (Figure 2). In addition, the antiviral effect of allicin on cytomegalovirus was also verified at the protein and mRNA level.
We found that allicin could reduce the level of IE2 protein in U87MG cells and further increase the expression of p53. Previous molecular and cytogenetic studies have provided compelling evidence that intracellular accumulation of p53 not only inhibits proliferation of cancer cells but also induces apoptosis.41,42 The HCMV IE2 protein plays an important role in p53 induction and transcriptional regulation of TP53 during HCMV infection. Part of the reason that IE2-86 inhibits the transcriptional activity of p53 may be that IE2-86 directly binds to the TP53 promoter and inhibits its transcription.29,43
We confirmed that allicin could reduce the inflammatory response of U87MG cells by downregulating IL-6 and IFN-β. Allicin could also inhibit cell proliferation. IL-6 may promote the proliferation, migration, and invasion of GBM by inducing STAT3 activation and downstream signaling.44,45 The decreased expression of IFN-β and IL-6 with allicin treatment suggested that allicin could inhibit HCMV activity and reduce the malignant degree of GBM.46,47 Allicin also inhibited the expression of IL-6 and IFN-β in macrophages, which could inhibit the protumor features of M2 macrophages, as well as their differentiation.48 Therefore, allicin may reduce immunosuppressive tumor-supporting macrophages in GBM, thereby inhibiting tumor proliferation.49
Another interesting finding in this study was that allicin increased the sensitivity of glioma cells to radiation. There is increasing evidence that radioresistance is the main cause of limited therapeutic effect in glioma patients. Moreover, a number of trials examining immunotherapy in glioma have shown promise.50 However, radiotherapy induces profound immunosuppression, which adds to that induced by the disease itself and by steroid administration,51 and therefore immunotherapies may not be beneficial in this context. The comet assay suggested that allicin had a radiosensitization effect on glioma cells. Therefore, a lower dose of radiotherapy might be able to elicit the antitumor immune responses. This radiosensitization may be due to allicin inhibiting the NF-κB signaling pathway in glioma cells.52
Allicin also inhibited proliferation in U87MG cells. This is likely because allicin reduced the level of IE2 protein and increased the expression of p53. Allicin also inhibited the expression of the inflammatory cytokines IL-6 and IFN-β in glioma cells. Additionally, allicin reduced the expression of IL-6 and IFN-β in macrophages. These results suggest that allicin can achieve an anti-tumor effect in glioma by blocking HCMV IE2, reducing the level of intracellular inflammatory factors (IFN-β and IL-6), and inhibiting macrophage infiltration. These specific targets may provide new ideas for future research in glioma treatment.
However, this study has several limitations. First, it was observed that allicin could reduce IE2 protein and IL-6 and IFN-β mRNA levels in HCMV-infected gliomas, thereby inhibiting the proliferation of glioma cells. However, the internal molecular mechanism was not further explored, and the possible mechanism was only clarified through existing studies. Second, only U87MG cells were used. In subsequent studies, more glioma cell lines, such as U-251 or GL-261, should be examined. Animal studies should also be performed to assess the efficacy of allicin in vivo.
Conclusion
In summary, this study confirmed that allicin can inhibit the viability of HCMV-infected U87MG glioma cells and inhibit cell proliferation by inhibiting the inflammatory response. Allicin also downregulated the inflammatory cytokines IL-6 and IFN-β. Therefore, allicin should be considered as a new therapeutic drug for GBM, and the importance of antiviral therapies in the treatment of GBM should be examined. The exact molecular mechanism of proliferation inhibition induced by allicin and the antitumor effect of allicin in vivo remain to be further investigated.
Abbreviations
HMCV, human cytomegalovirus; GBM, glioblastoma multiforme; IE, immediately-early; IL, interleukin; IFN, interferon; PBS, phosphate-buffered saline; ELISA, enzyme-linked immunosorbent assay; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Ethics Approval
All experiments were approved by the ethics committee of The Second Affiliated Hospital of Wenzhou Medical University.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kelly K, Kirkwood JM, Kapp DS. Glioblastoma multiforme: pathology, natural history and treatment. Cancer Treat Rev. 1984;11(1):1–26. doi:10.1016/0305-7372(84)90014-8
2. Kala M, Srámek V, Houdek M, Vaverka M, Zmrzlík P. Zkusenosti s lécbou multiformního glioblastomu. [Treatment of glioblastoma multiforme]. Cas Lek Cesk. 1993;132(21):653–656.
3. Holland EC. Glioblastoma multiforme: the terminator. Proc Natl Acad Sci U S A. 2000;97(12):6242–6244. doi:10.1073/pnas.97.12.6242
4. Van Meir E, Hadjipanayis C, Norden A, Shu H, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. doi:10.3322/caac.20069
5. Cobbs C, Soroceanu L, Denham S, Zhang W, Kraus MH. Modulation of oncogenic phenotype in human glioma cells by cytomegalovirus IE1-mediated mitogenicity. Cancer Res. 2008;68(3):724–730. doi:10.1158/0008-5472.CAN-07-2291
6. Cobbs C, Harkins L, Samanta M, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62(12):3347–3350.
7. Da Mitchell XW, Schmittling R, et al. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro-Oncology. 2008;10(1):10–18. doi:10.1215/15228517-2007-035
8. Luo M, Hannemann H, Kulkarni A, Schwartz P, O’dowd JM, Fortunato EA. Human cytomegalovirus infection causes premature and abnormal differentiation of human neural progenitor cells. J Virol. 2010;84(7):3528–3541. doi:10.1128/JVI.02161-09
9. Odeberg J, Wolmer N, Falci S, et al. Late human cytomegalovirus (HCMV) proteins inhibit differentiation of human neural precursor cells into astrocytes. J Neurosci Res. 2007;85(3):583–593. doi:10.1002/jnr.21144
10. Fornara O, Bartek J
11. Cinatl J, Scholz M, Kotchetkov R, Vogel JU, Doerr HW. Molecular mechanisms of the modulatory effects of HCMV infection in tumor cell biology. Trends Mol Med. 2004;10(1):19–23. doi:10.1016/j.molmed.2003.11.002
12. Hu M, Wang B, Qian D, et al. Human cytomegalovirus immediate-early protein promotes survival of glioma cells through interacting and acetylating ATF5. Oncotarget. 2017;8(19):32157–32170. doi:10.18632/oncotarget.17150
13. Schwartz R, Helmich B, Spector DH. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J Virol. 1996;70(10):6955–6966. doi:10.1128/JVI.70.10.6955-6966.1996
14. Huang R, Qian D, Hu M, et al. Association between human cytomegalovirus infection and histone acetylation level in various histological types of glioma. Oncol Lett. 2015;10(5):2812–2820.
15. Agarwal K. Therapeutic actions of garlic constituents. Med Res Rev. 1996;16(1):111–124. doi:10.1002/(SICI)1098-1128(199601)16:1<111::AID-MED4>3.0.CO;2-5
16. Butt M, Sultan M, Butt MS, Iqbal J. Garlic: nature’s protection against physiological threats. Crit Rev Food Sci. 2009;49(6):538–551. doi:10.1080/10408390802145344
17. Lissiman E, Bhasale A, Cohen M. Garlic for the common cold. Cochrane Database Syst Rev. 2014;undefined(11):CD006206.
18. Zou X, Liang J, Sun J, et al. Allicin sensitizes hepatocellular cancer cells to anti-tumor activity of 5-fluorouracil through ROS-mediated mitochondrial pathway. J Pharmacol Sci. 2016;131(4):233–240. doi:10.1016/j.jphs.2016.04.017
19. Chu Y, Ho C, Chung J, Raghu R, Lo YC, Sheen LY. Allicin induces anti-human liver cancer cells through the p53 gene modulating apoptosis and autophagy. J Agri. 2013;61(41):9839–9848.
20. Hirsch K, Danilenko M, Giat J, et al. Effect of purified allicin, the major ingredient of freshly crushed garlic, on cancer cell proliferation. Nutri Cancer. 2000;38(2):245–254. doi:10.1207/S15327914NC382_14
21. Li C, Jing H, Ma G, Liang P. Allicin induces apoptosis through activation of both intrinsic and extrinsic pathways in glioma cells. Mol Med Rep. 2018;17(4):5976–5981. doi:10.3892/mmr.2018.8552
22. Cha J, Choi Y, Cha S, Choi CH, Cho WH. Allicin inhibits cell growth and induces apoptosis in U87MG human glioblastoma cells through an ERK-dependent pathway. Oncol Rep. 2012;28(1):41–48. doi:10.3892/or.2012.1772
23. Silginer M, Nagy S, Happold C, Schneider H, Weller M, Roth P. Autocrine activation of the IFN signaling pathway may promote immune escape in glioblastoma. Neuro-Oncology. 2017;19(10):1338–1349. doi:10.1093/neuonc/nox051
24. Haller O, Kochs G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J Interferon Cytokine Res. 2011;31(1):79–87. doi:10.1089/jir.2010.0076
25. Browne E, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75(24):12319–12330. doi:10.1128/JVI.75.24.12319-12330.2001
26. Lagneaux L, Delforge A, Snoeck R, et al. Human cytomegalovirus increases constitutive production of interleukin-6 and leukemia inhibitory factor by bone marrow stromal cells. Blood. 1996;87(1):59–66. doi:10.1182/blood.V87.1.59.59
27. Rossini G, Cerboni C, Santoni A, et al. Interplay between human cytomegalovirus and intrinsic/innate host responses: a complex bidirectional relationship. Mediators Inflamm. 2012;2012(undefined):607276. doi:10.1155/2012/607276
28. Hori T, Sasayama T, Tanaka K, et al. Tumor-associated macrophage related interleukin-6 in cerebrospinal fluid as a prognostic marker for glioblastoma. J Clin Neurosci. 2019;68(undefined):281–289. doi:10.1016/j.jocn.2019.07.020
29. Speir E, Modali R, Huang E, et al. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science (New York, NY). 1994;265(5170):391–394. doi:10.1126/science.8023160
30. Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15(7):422–442. doi:10.1038/s41571-018-0003-5
31. Shea A, Harish V, Afzal Z, et al. MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics. Cancer Med. 2016;5(8):1917–1946.
32. Xing Y, Wang Y, Wang S, et al. Human cytomegalovirus infection contributes to glioma disease progression via upregulating endocan expression. Transl Res. 2016;177:113–126.
33. Rahbar A, Cederarv M, Wolmer-Solberg N, et al. Enhanced neutrophil activity is associated with shorter time to tumor progression in glioblastoma patients. Oncoimmunology. 2016;5(2):e1075693. doi:10.1080/2162402X.2015.1075693
34. Chamberlain M. Treatment options for glioblastoma. Neurosurg Focus. 2006;20(4):E19. doi:10.3171/foc.2006.20.4.12
35. Weber N, Andersen D, North J, Murray B, Lawson LD, Hughes BG. In vitro virucidal effects of Allium sativum (garlic) extract and compounds. Planta Med. 1992;58(5):417–423. doi:10.1055/s-2006-961504
36. Ruddock P, Liao M, Foster B, Lawson L, Arnason JT, Dillon JA. Garlic natural health products exhibit variable constituent levels and antimicrobial activity against Neisseria gonorrhoeae, Staphylococcus aureus and Enterococcus faecalis. Phytother Res. 2005;19(4):327–334. doi:10.1002/ptr.1667
37. Tsai Y, Cole L, Davis L, Lockwood S, Simmons V, Wild GC, Antiviral properties of garlic: in vitro effects on influenza B, herpes simplex and coxsackie viruses. Planta Med. 1985;5:460–461. doi:10.1055/s-2007-969553
38. Lissiman E, Bhasale Al CM. Garlic for the common cold. Cochrane Database Syst Rev. 2012;3:CD006206.
39. Patya M, Zahalka M, Vanichkin A, et al. Allicin stimulates lymphocytes and elicits an antitumor effect: a possible role of p21ras. Int Immunol. 2004;16(2):275–281. doi:10.1093/intimm/dxh038
40. Kyung K. Antimicrobial properties of allium species. Curr Opin Biotechnol. 2012;23(2):142–147. doi:10.1016/j.copbio.2011.08.004
41. Baker S, Markowitz S, Fearon E, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science (New YorkN Y). 1990;249(4971):912–915. doi:10.1126/science.2144057
42. Isaacs W, Carter BS, Ewing CM. Wild-type p53 suppresses growth of human prostate cancer cells containing mutant p53 alleles. Cancer Res. 1991;51(17):4716–4720.
43. Tsai H, Kou G, Chen S, Wu CW, Lin YS. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J Biol Chem. 1996;271(7):3534–3540. doi:10.1074/jbc.271.7.3534
44. Chen Z, Yuan S, Li K, et al. Doxorubicin-polyglycerol-nanodiamond conjugates disrupt STAT3/IL-6-mediated reciprocal activation loop between glioblastoma cells and astrocytes. J Control Release. 2020;320:469–483. doi:10.1016/j.jconrel.2020.01.044
45. Cutone A, Colella B, Pagliaro A, et al. Native and iron-saturated bovine lactoferrin differently hinder migration in a model of human glioblastoma by reverting epithelial-to-mesenchymal transition-like process and inhibiting interleukin-6/STAT3 axis. Cell Signal. 2020;65:109461. doi:10.1016/j.cellsig.2019.109461
46. Huang R, Qian D, Hu M, et al. Association between human cytomegalovirus infection and histone acetylation level in various histological types of glioma. Oncol Lett. 2015;10(5):2812–2820.
47. Pautasso S, Galitska G, Dell’oste V, et al. Strategy of human cytomegalovirus to escape interferon beta-induced APOBEC3G editing activity. J Virol. 2018;92(19).
48. Sousa S, Brion R, Lintunen M, et al. Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res. 2015;17:101. doi:10.1186/s13058-015-0621-0
49. Glass R, Synowitz M. CNS macrophages and peripheral myeloid cells in brain tumours. Acta Neuropathol. 2014;128(3):347–362. doi:10.1007/s00401-014-1274-2
50. Dutoit V, Migliorini D, Dietrich PY, Walker PR. Immunotherapy of malignant tumors in the brain: how different from other sites? Front Oncol. 2016;6:256. doi:10.3389/fonc.2016.00256
51. Pitter K, Tamagno I, Alikhanyan K, et al. Corticosteroids compromise survival in glioblastoma. Brain. 2016;139:1458–1471. doi:10.1093/brain/aww046
52. Huang W, Wu S, Xu S, et al. Allicin enhances the radiosensitivity of colorectal cancer cells via inhibition of NF-κB signaling pathway. J Food Sci. 2020;85(6):1924–1931. doi:10.1111/1750-3841.15156
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.