Back to Journals » Cancer Management and Research » Volume 12
Adipose Mesenchymal Stem Cell-Derived Exosomal microRNA-1236 Reduces Resistance of Breast Cancer Cells to Cisplatin by Suppressing SLC9A1 and the Wnt/β-Catenin Signaling
Authors Jia Z, Zhu H, Sun H, Hua Y, Zhang G, Jiang J, Wang X
Received 1 July 2020
Accepted for publication 21 August 2020
Published 22 September 2020 Volume 2020:12 Pages 8733—8744
DOI https://doi.org/10.2147/CMAR.S270200
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Zhongming Jia,1,* Huamin Zhu,2,* Hongguang Sun,1 Yitong Hua,1 Guoqiang Zhang,1 Jingru Jiang,1 Xiaohong Wang1
1Department of Thyroid and Breast Surgery, Affiliated Hospital of Binzhou Medical University, Binzhou 256603, Shandong, People’s Republic of China; 2Department of Medical Ultrasonics, Affiliated Hospital of Binzhou Medical University, Binzhou 256603, Shandong, People’s Republic of China
*These authors contributed equally to this work.
Correspondence: Zhongming Jia Tel +86-543-325779
Email [email protected]
Background: Emerging evidence has noted the versatile functions of mesenchymal stem cell-derived exosomes (MSC-Exos) in cancer control. This work aims to probe to function of adipose MSC-Exos (adMSC-Exos) in drug-resistance of breast cancer (BC) cells to cisplatin (DDP) and the molecules involved.
Methods: Parental and DDP-resistant BC cell lines MCF-7 and MDA-MB-231 were used. All cells were pre-treated with adMSC-Exos. Then, the viability and apoptosis of cells after DDP treatment were determined. Differentially expressed miRNAs after adMSC-exo treatment were screened out. Rescue experiments were conducted by pre-transfecting miR-1236 inhibitor into adMSCs, and the role of miR-1236 in DDP sensitivity was determined. Targeting mRNAs of miR-1236 were predicted by bioinformatics analysis. Altered SLC9A1 expression was administrated to evaluate its function in DDP resistance.
Results: The adMSC-Exos notably increased the sensitivity of either parental or DDP-resistant BC cells to DDP. SLC9A1 was notably highly expressed in DDP-resistant cells but inhibited following adMSC-exo administration. Importantly, miR-1236, which could directly bind to SLC9A1 and suppress its expression, was confirmed as an enriched miRNA in adMSC-Exos. Either inhibition of miR-1236 or upregulation of SLC9A1 blocked the pro-sensitize roles of adMSC-Exos. In addition, the Wnt/β-catenin pathway activity was suppressed by adMSC-Exos but recovered by SLC9A1.
Conclusion: This study evidenced that adMSC-Exos carry miR-1236 to increase sensitivity of BC cells to DDP with the involvement of SLC9A1 downregulation and Wnt/β-catenin inactivation. This finding may offer novel insights into treatment for drug-resistant BC.
Keywords: breast cancer, cisplatin, resistance, adipose mesenchymal stem cell-derived exosomes, microRNA-1236, SLC9A1
Introduction
Breast cancer (BC) is the most frequent neoplastic disease and the leading contributor to cancer death among females across the globe.1 It is featured with the progressive accumulation of mutations which result in the uncontrolled development of breast tissue cells.2 The major risk factors of BC comprise a family history, female gender, age, alcohol, and oral contraceptive.3 The mortality of BC has seen a stable decline in the past decades, and an early diagnosis is crucial for a better outcome of patients.4 Though surgical resection is the primary treatment strategy for BC at any disease stage, neoadjuvant chemotherapy, usually administrated prior to surgery, is frequently applied to decrease tumor size and suppress micrometastases.5 Cisplatin (DDP) is one of the commonest chemotherapy regimens for several cancers including BC, especially for triple-negative BC that shows insufficient response to anti-HER-2 or endocrine therapies.6 However, though patients show positive response to DDP treatment in the initial treatment, drug resistance and considerable side effects are remaining as major challenges.7 Developing novel efficacious treatments for BC and/or new strategies to overcome the chemo-resistance of BC patients has been a major issue in this field.
Exosomes are the most widely studied extracellular vehicles with 30–150 nm in diameter and implicated in intercellular communication, carrying multiple molecules including proteins, lipids, nucleic acids [DNA, mRNA, and microRNAs (miRNAs)] and specific cell-surface markers.8–10 Exosomes derived from tumor and stem cells have been involved in almost all stages in the progression of cancer and the therapy resistance as well as prognosis.11 Mesenchymal stem cells (MSCs) are one of the most ideal cell therapy tools, and the adipose-derived MSCs (adMSCs) are recognized as an abundant and potent tool in cell therapy.12 In addition, the adMSC-derived exosomes (adMSC-Exos) play versatile functions in cellular behaviors such as proliferation, migration, apoptosis, have become multifunctional frontiers in contemporary medicine.12 A study by Salha et al suggested that adMSCs are likely to migrate towards carcinoma sights in BC by platelet-derived growth factors,13 while a previous study noted that adMSC-Exos exert key anti-proliferation functions in ovarian cancer cells through the cargo miRNAs.14 But the exact roles of adMSC-Exos in BC progression and chemoresistance remain largely unknown.
Importantly, our bioinformatics analysis using a Gene Expression Omnibus (GEO) chip identified SLC9A1 as a significantly upregulated gene in DPP-resistant BC cells. SLC9A1, also termed Na/H exchanger 1 (NHE1), is a highly conserved plasma membrane protein that plays crucial roles in cell migration, proliferation and death, and it was noted to trigger the sensitivity of ΔNErbB2-expressing MCF-7 cells to DDP.15 Intriguingly, our study found that miR-1236, an enriched miRNA in adMSC-Exos, was found as a upstream regulator of SLC9A1. Importantly, a recent study noted that downregulation of miR-1236 by long non-coding RNA NNT-AS1 increased the resistance of lung cancer cells to DDP.16 Thereby, this study hypothesized that adMSC-Exos carry miR-1236 to suppress SLC9A1 expression, therefore inhibiting BC progression and the drug resistance of BC cells.
Materials and Methods
Collection and Identification of adMSC-Derived Exosomes
The adMSCs from ATCC (Manassas, VA, USA) were cultivated in DMEM supplementing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (all provided by Gibco Company, Grand Island, NY, USA). The cells were labeled with mouse anti-human CD90-fluorescein isothiocyanate (FITC), CD29-FITC, CD105-FITC, CD34-PE, CD44-PE, CD73-FITC, CD45-947 and HLA-DR (all purchased from Cell Signaling Technology, Beverly, MA, USA), and the positive labeling was determined by flow cytometry.
The osteogenic and adipogenic differentiation abilities of adMSCs were examined using the OriCellTM osteogenic differentiation and adipogenic differentiation induction kits (Cyagen Biosciences Inc, CA, USA) in accordance with the kit’s instructions.
Once reaching an 80% confluence, the well-growing passage 3 cells were incubated in serum-free medium for 24 h at 37°C, after which the supernatant was collected, named conditioned medium (CM). The CM was successively centrifuged at 300 g for 10 min, 2000 g for 30 min, and 10,000 g for 1 h, and then filtered using a 0.22 μm filter. The cell debris and large-size particles were discarded. After protein concentration determination using a bicinchoninic acid (BCA) kit (Solarbio Science & Technology Co., Ltd., Beijing, China), the extracted exosomes were preserved at −80°C.17
The exosome resuspension was prepared and dripped on the sealing film for transmission electron microscope (TEM) observation. To protect the exosomes from destruction, a copper mesh was used and placed on the drops of resuspension for 20 min of absorption. Then, the remaining water was absorbed by filter papers, after which the copper mesh was further placed on glutaraldehyde drops for 5 min, dried, and then placed on hydrogen peroxide solution drops for 2 min. This process was repeated 7 times. After absorption of the redundant water, the copper mesh with exosomes were further placed on uranyl acetate drops for 10 min, dried, and then on 1% methylcellulose drops for 5 min (all processes were performed on ice). After being air-dried, the exosomes were observed under an 80 kV TEM. The expression of exosomal marker proteins CD9, CD81, CD63 and heat shock protein 70 (HSP 70) (all from Abcam Inc., Cambridge, MA, USA) was evaluated by Western blot assay.
Western Blot Analysis
Cells were lysed in lysis buffer (BC3711, Solarbio), and then the concentration was determined by BCA. An equal volume of protein (40 µg) was separated on 8% SDS-PAGE and transferred onto PVDF membranes (Thermo Fisher Scientific, Rockford, IL, USA). After blocked by 5% skimmed milk (Solarbio), the membranes were cultured with primary antibodies at 4°C for 16 h, and then with secondary antibody for at 20°C for 2 h. The immunoblots were developed by Western ECL substrate (PIERCE), and the gray value was measured by a Gel-Pro-Analyzer Software.
Construction of DDP-Resistant BC Cell Lines
MCF-7 and MDA-MB-231 cells were purchased from ATCC. In brief, the parental BC cells (defined MCF-7/Par and MDA-MB-231/Par) were continuously exposed to an ascending series of DDP (0.1~10 μM, Sigma-Aldrich, MO, USA) to induce DDP-resistant cells (MCF-7/DDP and MDA-MB-231/DDP). All cells were maintained in 10% FBS-supplemented DMEM containing 1% penicillin-streptomycin for further use.
Determination of the 50% Concentration of Inhibition (IC50) Value of DDP in BC Cells
The IC50 value of DDP in BC cells was evaluated by the CellTiter-Glo®(CTG) assay. The cells were seeded in 96-well plates (5×103 cells/well), and a CellTiter-Glo® kit (Promega Corporation, Wisconsin, USA) was utilized to measure cell viability. In brief, the viability of cells was measured according to the quantification of the detected Adenosine Triphosphate, which is an important biomarker for cells with highly active metabolism.
Cell Viability Detection
Cells were sorted into 6-well plates for two weeks of cultivation, after which the colonies were fixed and stained. The number of cell colonies (≥50 cells) was recorded. For 5-ethynyl-2ʹ-deoxyuridine (EdU) labeling assay, the BC cells were seeded at 5 × 103 cells/well in 96-well plates containing 50 mM EdU (RiboBio, Guangzhou, China). Twelve hours later, the cells were immobilized in 4% paraformaldehyde and incubated in 0.5% Triton X-100 for 10 min. Afterwards, the cells were incubated with 100 µL Apollo 567 for 30 min. The nuclei were stained with Hoechst 33,342, and the EDU-positive cells were counted under a TCS SP8 confocal microscope (Leica, Bannockburn, IL, USA).
Lactate Dehydrogenase (LDH) Measurement
The sensitivity of NC cells to DDP was further evaluated through the release of LDH in cells. In brief, after treatment, the cells were centrifuged at 2000g for 20 min, and then the LDH release in cells was determined using a LDH kit (Abcam) as per the kit’s instructions. The OD value was determined at a wavelength of at 490 nm.
Caspase-3 Activity Detection
A total of 5 × 104 cells were sorted into a 10-cm culture dish for 24 h of growing. Another 24 h after adMSC-exo and DDP treatment, the DMEM was replaced by serum-free medium. After 24 h, the caspase-3 activity in cells was determined by a Caspase-3 analysis kit (ab39401, Abcam).
Flow Cytometry
After transfection, the cells were detached in trypsin and collected. A total of 106 cells were resuspended in Binding Buffer and then administrated with Annexin V-FITC (5 μL) and PI solution (10 μL) for 15 min of reaction avoiding light exposure. Then, the cell apoptosis was analyzed on a flow cytometer with a C6 instrument (BD Biosciences, NJ, USA).
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
TRIzol Reagents (Thermo Fisher) were used to extract total RNA from cells. Next, the corresponding cDNA was obtained using a Reverse Transcription Kit (Takara Biotechnology Ltd., Dalian, China). Thereafter, a real-time qPCR was performed on a Bio-Rad CFX96 System using a SYBR Green Real-Time PCR Kit (Takara). The primers are listed in Table 1 with GAPDH and U6 set as internal references, respectively. Relative gene expression was measured using the 2−∆∆Ct method.
 |
Table 1 Primer Sequences for RT-qPCR |
Enzyme-Linked Immunosorbent Assay (ELISA)
Protein levels of Wnt/β-catenin signaling pathway-related factors Wnt1, β-catenin and CCND1 in cells were determined by ELISA kits in strict accordance with the manufacturer’s instructions (Abcam).
Luciferase Assay
The putative binding site between miR-1236 and SLC9A1 3ʹUTR was first predicted on TargetScan (http://www.targetscan.org/). Then, the SLC9A1 3ʹUTR containing the binding sequence with miR-1236 was inserted into the pmirGLO vectors (Promega) to construct SLC9A1 wild-type (WT) vectors (SLC9A1-WT), and the mutant type (MT) vectors (SLC9A1-MT) on the basis of the mutant binding sequences were constructed as well. Next, the WT and MT vectors were co-transfected with miR-1236 mimic or NC mimic in 293T cells (D0010, Solarbio) in 6-well plates. Cells were harvested 48 h after transfection, and a Dual-Luciferase Reporter Gene System (Promega) was utilized to evaluate the luciferase activity in cells.
Statistical Analysis
SPSS 21.0 (IBM Corp. Armonk, NY, USA) was applied for data analysis. Kolmogorov-Smirnov was utilized to check whether the data were in normal distribution. Data were expressed as mean ± standard deviation (SD). Data were compared by the t-test (two groups) and one-way or two-way analysis of variance (ANOVA, over 3 groups) with Tukey’s multiple comparisons test for post hoc check. p < 0.05 represents statistically significant difference.
Results
Identification of the adMSCs and the Exosomes
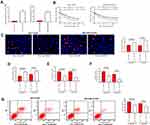
First, according to the flow cytometry, we identified positive expression of the surface biomarkers CD29, CD44, CD73, CD90 and CD105 while negative expression of CD14, CD34, HLA-DR and CD45 on the ad-MSCs (Figure 1A), indicating the separated and purified adMSCs in this research were in line with the current standard definition of MSCs. In addition, the oil red O and alizarin red staining results suggested that these adMSCs were able to differentiate into adipogenic cells and osteoblasts, respectively (Figure 1B–D). Next, the particles derived from adMSCs were observed under a TEM, under which the particles were in oval or cup-shape at a diameter of approximately 100 nm (Figure 1E). According to the guidelines in Minimal Information for Studies of Extracellular Vesicles 2018,18 we determined the exosome marker proteins CD9, CD63, CD81 and HSP70 as well as the cell marker protein GAPDH in the exosomes and cell lysates by Western blot assay. It was found that the expression levels of CD9, CD63, CD81 and HSP70 were significantly higher in exosomes than those in cell lysates, and only a poor level of GAPDH was found (Figure 1F), indicating the extracted particles were adMSC-Exos. The exosomes were diluted to 20 μg/mL for further use.
adMSC-Exos Decrease DDP Resistance of BC Cells
The drug-sensitive MCF-7/Par and MDA-MB-231/Par cells as well as the drug-resistant MCF-7-DDP and MDA-MB-231-DDP cells were treated with adMSC-Exos, after which the IC50 value of DDP in these cell lines was determined. Importantly, it was found that the resistance of both parental and drug-resistant cells to DDP was notably decreased following adMSC-exo treatment (Figure 2A). In addition, the EdU-labeling and colony formation assays suggested that the number of proliferative cells was notably declined after adMSC-exo and 5 μmol/mL or 10 μmol/mL DDP treatment (Figure 2B and C). The apoptosis of both parental and DDP-resistant cells was determined by flow cytometry. The results showed that either exposure to 5 μmol/mL or 10 μmol/mL DDP, the apoptosis rate of parental and drug-resistant cells was notably increased after adMSC-exo treatment (Figure 2D). Moreover, a LDH kit was used to determine the LDH secretion in cells after DDP exposure. It was found that the secretion of LDH in both cell lines was increased after adMSC-exo treatment, indicating an increase in toxicity of DDP in both cell lines (Figure 2E). In addition, the caspase-3 activity in cell lines was increased following adMSC-exo treatment as well (Figure 2F).
adMSC-Exos Increases Sensitivity of BC Cells to DDP Through Conveying miR-1236
To investigate the possible molecules responsible for the mediation by adMSC-Exos, we screened out the differentially expressed genes in a GEO chip GSE77515 containing the gene data of DDP-resistant MDA-MB-231 cells. A total of 339 differentially expressed genes were selected under the screening criteria |logFC| > 3 and adj p < 0.05 (Figure 3A). A heatmap for the top 30 genes is presented in Figure 3B. Next, the expression levels of the top 10 genes were measured by RT-qPCR, which suggested SLC9A1 owned the greatest differential expression in the MCF-7/DDP and MDA-MB-231/DDP cells (Figure 3C). Importantly, it was found that the mRNA expression of SLC9A1 in cells was notably decreased after adMSC-exo administration (Figure 3D). Thereby, we speculated that there might be a specific cargo carried by the exosomes specifically suppressing SLC9A1 expression. The enriched miRNAs in adMSC-Exos and the upstream regulator miRNAs of SLC9A1 were predicted on the bioinformatics systems EvmiRNA (http://bioinfo.life.hust.edu.cn/EVmiRNA#!/) and TargetScan, respectively, and miR-1236 was found to be intersected (Figure 3E). Next, a luciferase assay validated that co-transfection of miR-1236 mimic and SLC9A1-WT vector led to a notable decrease in the luciferase activity in 293T cells (Figure 3F), implicating that miR-1236 could directly bind to SLC9A1. Intriguingly, the RT-qPCR results found a poor expression profiling in MCF-7/DDP and MDA-MB-231/DDP cells, while further administration of adMSC-Exos led to increased miR-1236 expression in parental and drug-resistant BC cell lines (Figure 3G).
Reduction of miR-1236 Blocks the Promoting Roles of adMSC-Exos in DDP Sensitivity of BC Cells
To confirm the involvement of miR-1236 in the above events, miR-1236 inhibitor was pre-transfected into the adMSCs and then the corresponding exosomes were collected and named Exo/miR-1236 inhibitor. The RT-qPCR results showed that miR-1236 expression in these exosomes was reduced (Figure 4A). Thereafter, the parental and DDP-resistant cells were treated with the Exo/miR-1236 inhibitor and then exposed to series concentration of DDP. It was found that the IC50 value of DDP in both cell lines was notably increased (Figure 4B). Again, the cells with Exo/miR-1236 inhibitor transfection were exposed to 5 μmol/mL or 10 μmol/mL DDP, respectively, after which we found the viability and proliferation activity of cells were significantly promoted (Figure 4C and D), while the apoptosis rate, LDH secretion, and caspase-3 activity in cells were decreased (Figure 4E–G).
Overexpression of SLC9A1 Blocked the Functions of adMSC-Exos in DDP Sensitivity of BC Cells
The role of SLC9A1 in adMSC-exo/miR-1236-mediated events was determined by rescue experiments as well. Overexpression of SLC9A1 was introduced in parental and drug-resistant BC cell lines, and the successful upregulation was checked by the RT-qPCR (Figure 5A). Further, the cells were treated with the adMSC-Exos and then exposed to different doses of DDP, after which we found the IC50 value of DDP in both cells was increased (Figure 5B). Again, the numbers of EdU-positive cells and colonies formed were increased upon SLC9A1 overexpression (Figure 5C and D), while the apoptosis rate, LDH production, and caspase-3 activity in cells were declined (Figure 5E–G).
SLC9A1 Activates the Wnt/β-Catenin Signaling
A previous study noted that SLC9A1 may activate the Wnt/β-catenin axis.19 Here, we measured the levels of Wnt/β-catenin marker proteins Wnt1, β-catenin and CCND1 in parental and DDP-resistant BC cells. Consequently, the drug-resistant cells presented an increase in the levels of these proteins. However, the activity of this signaling was inhibited by the adMSC-Exos, but further recovered in the presence of miR-1236 inhibition or SLC9A1 overexpression (Figure 6A and B).
Discussion
The chemoresistance is a dominant barrier in the effectiveness of patients who receive chemotherapeutic intervention, and thus a contributor to cancer progression and morbidity.20 The MSCs and their cellular vehicles have been increasingly recognized as novel promising therapeutic options for BC treatment.21 In this paper, we identified an important function of adMSC-Exos in the reduction of DDP resistance of BC MCF-7 and MDA-MB-23 cells with the crucial involvement of miR-1236 as well as the suppression of SLC9A1 and the Wnt/β-catenin axis.
Recent studies have witnessed exosomes from different MSC sources presenting key mediatory functions in cancer cell behaviors from proliferation and migration to chemo and radio resistance.22–26 Here, our study explored the correlation between adMSC-Exos and the DDP resistance of BC cells. After identification of several MSC-specific biomarkers including CD63, CD9 and CD81, and HSP70,27,28 the collected adMSC-Exos were administrated into parental and DDP-resistant MCF-7 and MDA-MB-23 cells. Next, we observed that the sensitivity of both parental and drug-resistant cells to DDP was notably increased, presenting as reductions in the number of EdU-positive cells and cell colonies while an increase in cell apoptosis. From a cellular perspective, the results that the secretion of LDH and caspase-3 in cells further confirmed an increased cell apoptosis in cells after adMSC-Exo treatment. Quite in line with our study, a recent study noted that miR-199a-modified adMSC-Exos improved the chemosensitivity of hepatocellular carcinoma cells through the mTOR pathway.29 We therefore explored the potential molecules involved in the adMSC-Exos-mediated DDP sensitivity in BC cells.
Based on the findings above, we determined the significantly differentially expressed genes using a GEO GSE77515 chip containing the data of DDP-resistant MDA-MB-231 cells. Correlating the screened out genes with the further RT-qPCR results, we identified SCL9A1 as the most upregulated gene in DDP-resistant BC cells. SCL9A1 has been reported as an oncogene in several cancers.30 Similar roles of SCL9A1 have been reported in BC. For instance, silencing of SCL9A1 by CIAPIN1 was responsible for the inhibition in cell migration, inhibition, and matrix metalloproteinase production in MDA-MB-231 cells.31 Likewise, KR-33,028, a potent SCL9A1 antagonist, was found to reduce the metastatic potential of triple-negative BC cells.32 In terms of chemoresistance, SCL9A1 has been reported to hold accountable for the resistance of chronic myeloid leukemia cells to imatinib treatment, and inhibition of SCL9A1 was found to reduce intracellular pH,33 while inhibition of pH is also a promising therapeutic strategy for BC treatment.34 Similarly, downregulation of SCL9A1 by secreted frizzled-related protein 1 was evidenced to increase the sensitivity of laryngeal carcinoma cells to DDP.35 As for in BC, SCL9A1 inhibition was also demonstrated to sensitize the drug-resistant BC cells to doxorubicin treatment both in cell and animal models.36 Importantly, in our present study, the mRNA expression of SCL9A1 in BC cell lines was notably decreased, indicating that inhibition of SCL9A1 was possibly responsible for adMSC-Exos-mediated DDP sensitivity in cells. Based on this, we further predicted the enriched miRNAs in adMSC-Exos and the upstream miRNAs of SLC9A1 through the bioinformatics systems EvmiRNA and TargetScan, respectively. miR-1236 was suggested to be intersected. To further validate the involvement of miR-1236, the miR-1236-suppressed adMSC-Exos were collected. It was found that inhibition of miR-1236 in exosomes led to a significant increase in mRNA and protein expression of SCL9A1 in cells. Correspondingly, the promoting effects of adMSC-Exos on DDP sensitivity of BC cell lines were blocked after miR-1236 silencing. Interestingly, miR-1236 has been documented to reduce DDP resistance of lung cancer cells.16,37 Here, we further evidenced the anti-DDP resistance roles of this miRNA in BC cells.
Interestingly, Wnt/β-catenin was reported as one of the activated pathways under SCL9A1 mediation.19 This pathway is an evolutionarily conserved network that exerts key roles during embryonic and tissue development, stemness maintenance, and metastasis and tumor progression, thus serving as a hallmark of many tumors including BC.38 Aberrant activation of the Wnt/β-catenin pathway led to DDP resistance in several cancer types such as lung cancer,39 gastric cancer,40 and BC as well.41 Importantly, we found that this signaling was activated in DDP-resistant BC cells and then inhibited by the adMSC-Exos, but it was further recovered upon miR-1236 inhibition or SLC9A1 overexpression.
Conclusion
Collectively, this study evidenced that adMSC-Exos carry miR-1236 to suppress SLC9A1 expression and inactivate the Wnt/β-catenin axis, therefore reducing the resistance of BC cells to DDP (Figure 7). This study may offer novel insights into extracellular vesicles-based interventions for chemo resistance control. However, the major limitation of this research was that all experiments were carried out in cells. We would like to conduct studies in animal models to verify the effects of adMSC-Exos on DDP resistance in vivo. We also hope more studies in this field will be performed to provide more understanding for BC control.
Acknowledgments
We thank Hanyu Biomed Center for the help in bioinformatic analysis.
Author Contributions
ZGJ and HMZ contributed to the study design and manuscript preparation. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Mendes PMV, Bezerra DLC, Dos Santos LR, et al. Magnesium in breast cancer: what is its influence on the progression of this disease? Biol Trace Elem Res. 2018;184(2):334–339. doi:10.1007/s12011-017-1207-8
3. Cuchra M, Mucha B, Markiewicz L, et al. The role of base excision repair in pathogenesis of breast cancer in the Polish population. Mol Carcinog. 2016;55(12):1899–1914. doi:10.1002/mc.22436
4. Cronin KA, Lake AJ, Scott S, et al. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer. 2018;124(13):2785–2800. doi:10.1002/cncr.31551
5. Dunne M, Dou YN, Drake DM, et al. Hyperthermia-mediated drug delivery induces biological effects at the tumor and molecular levels that improve cisplatin efficacy in triple negative breast cancer. J Control Release. 2018;282:35–45. doi:10.1016/j.jconrel.2018.04.029
6. Wang Y, Li S, Zhu L, et al. Letrozole improves the sensitivity of breast cancer cells overexpressing aromatase to cisplatin via down-regulation of FEN1. Clin Transl Oncol. 2019;21(8):1026–1033. doi:10.1007/s12094-018-02019-1
7. Islam SS, Al-Sharif I, Sultan A, Al-Mazrou A, Remmal A, Aboussekhra A. Eugenol potentiates cisplatin anti-cancer activity through inhibition of ALDH-positive breast cancer stem cells and the NF-kappaB signaling pathway. Mol Carcinog. 2018;57(3):333–346. doi:10.1002/mc.22758
8. Canas JA, Sastre B, Rodrigo-Munoz JM, Del Pozo V. Exosomes: a new approach to asthma pathology. Clin Chim Acta. 2019;495:139–147. doi:10.1016/j.cca.2019.04.055
9. Lim W, Kim HS. Exosomes as therapeutic vehicles for cancer. J Tissue Eng Regen Med. 2019;16(3):213–223. doi:10.1007/s13770-019-00190-2
10. Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer. 2019;1871(2):455–468. doi:10.1016/j.bbcan.2019.04.004
11. Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol Cancer. 2019;18(1):32. doi:10.1186/s12943-019-0975-5
12. Hong P, Yang H, Wu Y, Li K, Tang Z. The functions and clinical application potential of exosomes derived from adipose mesenchymal stem cells: a comprehensive review. Stem Cell Res Ther. 2019;10(1):242. doi:10.1186/s13287-019-1358-y
13. Salha S, Gehmert S, Brebant V, et al. PDGF regulated migration of mesenchymal stem cells towards malignancy acts via the PI3K signaling pathway. Clin Hemorheol Microcirc. 2018;70(4):543–551. doi:10.3233/CH-189319
14. Reza A, Choi YJ, Yasuda H, Kim JH. Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci Rep. 2016;6:38498. doi:10.1038/srep38498
15. Lauritzen G, Jensen MB, Boedtkjer E, et al. NBCn1 and NHE1 expression and activity in DeltaNErbB2 receptor-expressing MCF-7 breast cancer cells: contributions to pHi regulation and chemotherapy resistance. Exp Cell Res. 2010;316(15):2538–2553. doi:10.1016/j.yexcr.2010.06.005
16. Wang H, Guo M, Ding D, Yang F, Chen Z. Long non-coding RNA NNT-AS1 contributes to cisplatin resistance via miR-1236-3p/ATG7 axis in lung cancer cells. Onco Targets Ther. 2020;13:3641–3652. doi:10.2147/OTT.S237576
17. Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278(22):20083–20090. doi:10.1074/jbc.M301642200
18. Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750.
19. Sun DI, Tasca A, Haas M, et al. Na+/H+ exchangers are required for the development and function of vertebrate mucociliary epithelia. Cells Tissues Organs. 2018;205(5–6):279–292. doi:10.1159/000492973
20. Wang X, He S, Gu Y, et al. Fatty acid receptor GPR120 promotes breast cancer chemoresistance by upregulating ABC transporters expression and fatty acid synthesis. EBioMedicine. 2019;40:251–262. doi:10.1016/j.ebiom.2018.12.037
21. Heidari R, Gholamian Dehkordi N, Mohseni R, Safaei M. Engineering mesenchymal stem cells: a novel therapeutic approach in breast cancer. J Drug Target. 2020;1–10.
22. de Araujo Farias V, O’Valle F, Serrano-Saenz S, et al. Exosomes derived from mesenchymal stem cells enhance radiotherapy-induced cell death in tumor and metastatic tumor foci. Mol Cancer. 2018;17(1):122. doi:10.1186/s12943-018-0867-0
23. Shojaei S, Hashemi SM, Ghanbarian H, Salehi M, Mohammadi-Yeganeh S. Effect of mesenchymal stem cells-derived exosomes on tumor microenvironment: tumor progression versus tumor suppression. J Cell Physiol. 2019;234(4):3394–3409. doi:10.1002/jcp.27326
24. Xie C, Du LY, Guo F, Li X, Cheng B. Exosomes derived from microRNA-101-3p-overexpressing human bone marrow mesenchymal stem cells suppress oral cancer cell proliferation, invasion, and migration. Mol Cell Biochem. 2019;458(1–2):11–26. doi:10.1007/s11010-019-03526-7
25. Xu Y, Shen L, Li F, Yang J, Wan X, Ouyang M. microRNA-16-5p-containing exosomes derived from bone marrow-derived mesenchymal stem cells inhibit proliferation, migration, and invasion, while promoting apoptosis of colorectal cancer cells by downregulating ITGA2. J Cell Physiol. 2019;234(11):21380–21394. doi:10.1002/jcp.28747
26. Zhang J, Song Q, Wu M, Zheng W. The emerging roles of exosomes in the chemoresistance of hepatocellular carcinoma. Curr Med Chem. 2020.
27. Ludwig N, Whiteside TL, Reichert TE. Challenges in exosome isolation and analysis in health and disease. Int J Mol Sci. 2019;20(19):4684. doi:10.3390/ijms20194684
28. Wang J, Yao Y, Wu J, Li G. Identification and analysis of exosomes secreted from macrophages extracted by different methods. Int J Clin Exp Pathol. 2015;8(6):6135–6142.
29. Lou G, Chen L, Xia C, et al. MiR-199a-modified exosomes from adipose tissue-derived mesenchymal stem cells improve hepatocellular carcinoma chemosensitivity through mTOR pathway. J Exp Clin Cancer Res. 2020;39(1):4. doi:10.1186/s13046-019-1512-5
30. Guan X, Luo L, Begum G, et al. Elevated Na/H exchanger 1 (SLC9A1) emerges as a marker for tumorigenesis and prognosis in gliomas. J Exp Clin Cancer Res. 2018;37(1):255. doi:10.1186/s13046-018-0923-z
31. Wang J, Xu H, Wang Q, et al. CIAPIN1 targets Na(+)/H(+) exchanger 1 to mediate MDA-MB-231 cells’ metastasis through regulation of MMPs via ERK1/2 signaling pathway. Exp Cell Res. 2015;333(1):60–72. doi:10.1016/j.yexcr.2015.02.012
32. Amith SR, Wilkinson JM, Fliegel L. KR-33028, a potent inhibitor of the Na(+)/H(+) exchanger NHE1, suppresses metastatic potential of triple-negative breast cancer cells. Biochem Pharmacol. 2016;118:31–39. doi:10.1016/j.bcp.2016.08.010
33. Ma D, Fang Q, Wang P, et al. Induction of heme oxygenase-1 by Na+-H+ exchanger 1 protein plays a crucial role in imatinib-resistant chronic myeloid leukemia cells. J Biol Chem. 2015;290(20):12558–12571. doi:10.1074/jbc.M114.626960
34. Meehan J, Ward C, Turnbull A, et al. Inhibition of pH regulation as a therapeutic strategy in hypoxic human breast cancer cells. Oncotarget. 2017;8(26):42857–42875. doi:10.18632/oncotarget.17143
35. Wu KM, Li ZQ, Yi WZ, et al. Restoration of secreted frizzled-related protein 1 suppresses growth and increases cisplatin sensitivity in laryngeal carcinoma cells by downregulating NHE1. Int J Clin Exp Pathol. 2017;10(8):8334–8343.
36. Chen Q, Liu Y, Zhu XL, Feng F, Yang H, Xu W. Increased NHE1 expression is targeted by specific inhibitor cariporide to sensitize resistant breast cancer cells to doxorubicin in vitro and in vivo. BMC Cancer. 2019;19(1):211. doi:10.1186/s12885-019-5397-7
37. Wang Z, Liu L, Guo X, Guo C, Wang W. microRNA-1236-3p regulates DDP resistance in lung cancer cells. Open Med (Wars). 2018;14:41–51. doi:10.1515/med-2019-0007
38. Prosperi JR, Goss KH. A Wnt-ow of opportunity: targeting the Wnt/beta-catenin pathway in breast cancer. Curr Drug Targets. 2010;11(9):1074–1088. doi:10.2174/138945010792006780
39. Zhang Q, Zhang B, Sun L, et al. MicroRNA-130b targets PTEN to induce resistance to cisplatin in lung cancer cells by activating Wnt/beta-catenin pathway. Cell Biochem Funct. 2018;36(4):194–202. doi:10.1002/cbf.3331
40. Cheng C, Qin Y, Zhi Q, Wang J, Qin C. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/beta-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol. 2018;107(Pt B):2620–2629. doi:10.1016/j.ijbiomac.2017.10.154
41. VanKlompenberg MK, Bedalov CO, Soto KF, Prosperi JR. APC selectively mediates response to chemotherapeutic agents in breast cancer. BMC Cancer. 2015;15:457. doi:10.1186/s12885-015-1456-x
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.