Back to Journals » Cancer Management and Research » Volume 12
Acetyl-11-keto-β-boswellic Acid Inhibits Precancerous Breast Lesion MCF-10AT Cells via Regulation of LINC00707/miR-206 that Reduces Estrogen Receptor-α
Authors Jiang X , Liu Y, Zhang G, Lin S, Yuan N , Wu J , Yan X, Ma Y, Ma M
Received 10 November 2019
Accepted for publication 20 February 2020
Published 27 March 2020 Volume 2020:12 Pages 2301—2314
DOI https://doi.org/10.2147/CMAR.S238051
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Xuefeng Jiang,1,* Yusheng Liu,1,* Guijuan Zhang,2,* Shujun Lin,1 Naijun Yuan,1 Jieyan Wu,1 Xianxin Yan,1 Yi Ma,3 Min Ma1,2
1College of Traditional Chinese Medicine of Jinan University, Guangzhou, People’s Republic of China; 2The First Affiliated Hospital of Jinan University, Guangzhou, People’s Republic of China; 3Institute of Biomedicine and Department of Cellular Biology, Jinan University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Min Ma
College of Traditional Chinese Medicine of Jinan University, 601 Huangpu Avenue West, Guangzhou, Guangdong 510632, People’s Republic of China
Tel +86 15876583624
Fax +86 20-85227173
Email [email protected]
Purpose: Acetyl-11-keto-β-boswellic acid (AKBA) has therapeutic effects on a range of diseases, including tumours. lncRNAs, as competing endogenous RNAs (ceRNAs), can interact with miRNAs to regulate the expression of target genes, which can affect the development of tumors. Here, we examined the effects of AKBA on breast precancerous lesions MCF-10AT cells.
Methods: The expression profiles of breast cancer (BC) tissue were collated from The Cancer Genome Atlas (TCGA), and the lncRNA-miRNA-mRNA ceRNA network was constructed. AKBA targets were predicted by network pharmacology. The expression of long intergenic nonprotein-coding RNA 707 (LINC00707), miR-206 and ER-α was determined by qRT-PCR. Cell viability, apoptosis and cycle were assessed by CCK-8 and flow cytometry. Protein levels were measured by Western blotting.
Results: A total of 3205 differentially expressed mRNAs, 104 miRNAs, and 605 lncRNAs were identified. The ceRNA network consisting of 9 lncRNAs, 15 miRNAs and 82 mRNAs was constructed. We found that LINC00707 was up-regulated and miR-206 was down-regulated in MCF-10AT cells. Transfected si-LINC00707 could inhibit cell proliferation, induce cell apoptosis and cycle arrest of MCF-10AT cells. In addition, network pharmacology predicted that AKBA may regulate the ESR1 in the treatment of BC. Our research demonstrated that AKBA could induce cell apoptosis and G1-phase arrest and inhibit ER-α expression via LINC00707/miR-206 in MCF-10AT cells.
Conclusion: AKBA inhibited MCF-10AT cells via regulation of LINC00707/miR-206 that reduces ER-α.
Keywords: breast precancerous lesion, acetyl-11-keto-β-boswellic acid, ceRNA, LINC00707, miR-206, ESR1
Introduction
BC is caused by mutation of multigene in mammary epithelial cells of the acini and ducts, resulting in normal mammary epithelium being transformed into tumorous epithelium through usual ductal hyperplasia (UDH)-atypical ductal epithelial hyperplasia (ADH)-ductal carcinoma in situ (DCIS)-invasive breast cancer (IBC).1 Atypical hyperplasia and carcinoma in situ may remain relatively stable for a long time before developing into invasive carcinoma, which are called precancerous lesion. According to the American cancer center 2018 estimated, there were 266,120 new cases of DCIS and 63,960 lobular carcinoma in situ (LCIS), respectively.2 And it alone was anticipated to account for 30% of all new cancer diagnoses in female. Therefore, the incidence of BC could be effectively decreased by blocking tumor progression.
Encyclopedia of DNA elements demonstrated that the mammalian genome are pervasively transcribed into many different complex families of RNA. In addition to a number of alternative transcriptional start sites, termination and splicing patterns, a complex collection of new antisense, intronic and intergenic transcripts was found.3 Moreover, most genomes (>98%) are transcribed into non-protein-coding RNAs, including lncRNAs, miRNAs, small interfering RNAs, small nuclear RNAs, and ribosomal RNAs. MiRNAs are a class of small ncRNAs with important regulatory roles, ~22 nt in length. They can posttranscriptionally regulate gene expression through binding to miRNA response elements (MREs) on their target transcripts. For example, miR-206 and miR-203 could promote tumor cell proliferation and stemness in BC by targeting mRNAs in vitro and in vivo.4,5 Although the function of some miRNAs has been well characterized to date, very little is known about the lncRNA counterpart of the transcriptome. LncRNAs are generally defined as RNA transcripts longer than 200 nucleotides. Recently, a new regulatory mechanism has been found that cross-talk between lncRNAs and mRNAs occurs by competing for shared MREs.6 LncRNAs can act as endogenous miRNA sponges to inhibit miRNA function, thereby impact the multiple targets of miRNAs. RNA transcripts sharing one or more MREs can actively regulate their respective expression levels by competing for a limited pool of miRNAs. For example, lncRNA H19 could mediate BC cell plasticity during EMT by differentially sponging miR-200b/c and let-7b.7
Natural products play a critical role in the discovery and the development of numerous drugs for the treatment of various types of deadly diseases including cancer.8,9 AKBA, a pentacyclic terpenoid, is the active component of frankincense and has been exploited for various medicinal applications, such as antibacterial, anti-inflammatory, analgesic, anti-oxidant.10,11 Recently, AKBA was also found to exhibit antitumor effects in human cell lines established from prostate cancer, breast cancer and glioblastoma.12–14 Csuk et al demonstrated that boswellic acid and its derivatives induce apoptosis in breast and cervical cancers cells.15 Liu indicated that AKBA inhibits the proliferation and cancer stem cell-like properties of docetaxel resistant prostate cancer cells in vitro and in vivo via blocking Akt and Stat3 signaling.16 However, the potential therapeutic effect and the underlying molecular mechanism of AKBA on breast tumor have yet to be elucidated.
In this study, we comprehensively integrated expression profiles, including data on mRNAs, miRNAs and lncRNAs of BC tissues and non-tumor tissues, and constructed the ceRNA network to describe the potential biological function in the development of BC. In addition, network pharmacology predicted that AKBA may regulate the ESR1 expression. Our research confirmed that AKBA can inhibit ER-α through LINC00707/miR-206 in breast precancerous lesions MCF-10AT cells. AKBA also induced MCF-10AT cell apoptosis and G1-phase arrest. These results would help to understand the mechanism of AKBA in preventing and curing BC.
Materials and Methods
Data Collection and Processing
The RNA expression data (level 3) were downloaded from TCGA data portal (https://portal.gdc.cancer.gov/). RNA-seq data (lncRNA and mRNA) from 113 non-tumor tissues and 1104 breast cancer tissues were included, as well as miRNA-seq data from 104 normal tissues and 1098 breast cancer tissues. This study was conducted in accordance with the publication guidelines provided by the TCGA (http://cancergenome.nih.gov/publications/publicationguidelines). Therefore, further approval of the ethics committee was not required.
Differential Expression Analysis
LncRNA and mRNA were defined and encoded according to gene labels from the Ensembl database (http://www.ensembl.org/index.html). We analyzed the differentially expressed lncRNA (DElncRNA), mRNA (DEmRNA), miRNA (DEmiRNA) using the edgeR package in software R, which is publicly available through Bioconductor (http://www.bioconductor.org/).17 False discovery rate (FDR)<0.01 and | LogFC |>1.5 were selected as the differentially expressed genes (DEGs) identification threshold. The ggplot2 package in R platform was used to visualize the heatmap and volcano plot.
Prediction of Target Genes and Construction of ceRNA Network
According to miRcode (http://www.mircode.org/) and miRBase (http://www.mirbase.org/), we analyzed the possible relationship between DElncRNA and DEmiRNA. Next, we used miRDB (http://mirdb.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and TargetScan (http://www.targetscan.org/mamm) to predict the miRNA–mRNA interactions. The target mRNAs were selected when the relationship between the DEmiRNA in miRDB, miRTarBase, and TargetScan is consistent with the predicted mRNA. Cytoscape v3.7.1 software was used to build an interactive and visual ceRNA network.
Functional Enrichment Analysis
In order to study the DEmRNA potential biological mechanism in ceRNA network, we used DAVID Bioinformatics Tool (https://david.ncifcrf.gov/) to identify the Gene Ontology (GO) annotation and Kyoto Encyclopedia Gene and genomes (KEGG) pathways, and with p values < 0.05 as the threshold of enrichment analysis. The GOplot package of R software was used to display the results of GO analysis.
PPI Network Analysis
DEmRNAs in the ceRNA network were uploaded to STRING (Version: 11.0, https://string-db.org/cgi/input.pl) to construct a protein–protein interaction (PPI) network. Visualization was carried out by Cytoscape 3.7.1. Meanwhile, cytoHhbba plug-in was used to identify highly interacting hub-gene clusters.
Target Prediction of AKBA
To identify the key sites, signaling pathways and biological processes involved in drug intervention, AKBA (PubChem CID: 17973666) was submitted to Bioinformatics Analysis Tool for Molecular mechanism of TCM (BATMAN-TCM, http://bionet.ncpsb.org/batman-tcm/).18 The predicted targets with scores ≥20 were presented. KEGG analysis was used to screen key targets and related signaling pathways. Meanwhile, disease enrichment analyses were performed based on disease-gene associations from Therapeutic Target Database (TTD, https://en.wikipedia.org/wiki/therapeutic-targets-database). Then, we constructed an ingredients-targets-diseases network to predict its efficiency on BC.
Cell Culture and Transfection
MCF10A and MCF-7 cell lines were purchased from the American Type Culture Collection (ATCC) and cultured according to manufacturer’s directions. MCF-10AT cell line was obtained from American Karmanos Cancer Institute (KCI). The human breast MCF-10A cell line originated from spontaneous immortalization of breast epithelial cells from a patient with fibrocystic disease. MCF-10AT cell derived from xenograft-passaged H-ras transfected MCF10A (MCF10A-ras) breast epithelial cells. MCF-7 cell line was luminal estrogen receptor-positive BC cell line. MCF-10AT cell was monolayer adherent cell. MCF-10A and MCF-10AT cells were maintained in DMEM/F12 (1:1) containing 5% horse serum, 20 ng/mL EGF, 10 μg/mL insulin, 50 μg/mL hydrocortisone. All cells were incubated in a humidified atmosphere of 5% CO2 at 37 °C.
LINC00707 siRNA (si-LINC00707), miR-206 mimic and inhibitor were transfected into cells using Lipofectamine 3000 (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol. Transfection efficiency was quantified by counting green fluorescent protein (GFP)-positive cells 24 hrs after transfection and found to be about 60–70%.
Cell Counting Kit 8 Assays
The cell viability was measured using CCK-8 assay (Dojindo Molecular Technologies, Tokyo, Japan). Cells were seeded in 96-well plates overnight. Then, the medium was replaced with the different concentrations of AKBA medium solution. After cultured for 24h, 10 μL of 5 mg/mL CCK-8 solution was added to each well for a further 2h incubation. Cell proliferation was measured at 450 nm using a microplate reader.
Annexin V/PI Staining Assay for Apoptosis
MCF-1A0T cells were collected and resuspended in binding buffer at a density of 1× 106 cells/mL. After staining the cells with Annexin V-FITC/propidium iodide (PI) (BD Biosciences, San Jose, CA, USA) for 15 min in the dark. The apoptotic cell death rate was examined using the flow cytometry.
Cell Cycle Analysis
The established cells were digested with 0.25% trypsin, washed 3 times with PBS buffer, and fixed with 70% alcohol at 4°C. Next, MCF-10AT cells were stained with 25μL PI (Vazyme, Nanjing, China) in the presence of 10μL RNase A at least for 30 min at 4°C. Flow cytometry was used to detect the red fluorescence at 488 nm excitation wavelength.
Quantitative Real-Time PCR
The RNAiso Plus (Takara, Japan) was used to obtain total RNAs. Then, the cDNA was synthesized from total RNA using QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). Subsequently, qRT-PCR was performed using SYBR Premix Ex Taq II (Takara, Japan) on Applied Biosystems 7900 Real-Time PCR System with the primers manifested in Table 1. Relative gene expression was calculated using the 2−ΔΔCt method.
 |
Table 1 Primers for Quantitative Real-Time PCR |
Western Blot Analysis
Total protein was extracted using the RIPA Lysis Buffer (Beyotime, China). Protein concentration was quantified by BCA protein assay (Beyotime, China). Equivalent amounts of protein were separated by 10% SDS-PAGE, and transferred onto PVDF membranes. After blocking the 5% nonfat milk solution for 1h, the membrane was incubated the respective primary antibodies (1:1000) overnight at 4°C. Subsequently, the membrane was probed with a goat anti-rabbit IgG secondary antibody at room temperature for 1h. Finally, the protein blots were visualized using ECL-Plus reagent (Millipore). The following antibodies were used in this study: ER-α, Bcl2, Bax, caspase-3, cleaved caspase-3, Cyclin D1, CDK4, CDK6, P27 and GAPDH (Cell Signaling Technology, Beverly, MA, USA).
Luciferase Reporter Assay
The wild-type (WT) or mutant (MUT) type of LINC00707 3′-untranslated region (3′-UTR) was synthesized and fused to a luciferase reporter vector psiCHECK-2 (Promega, Madison, WI, USA). MCF-10AT cells were co-transfected with reporter vector and miR-206 mimics or negative control. After transfection for 48 h, the relative luciferase activity was measured using a Dual-Luciferase Reporter Assay System according to the manufacturer’s instructions.
Statistical Analysis
The experimental data were analyzed with Student’s t-test and One-Way ANOVA Test using SPSS 17.0 software. All data were expressed as mean ± SD of at least three experiments. P<0.05 was considered statistically significant.
Results
Identify DEGs in BC
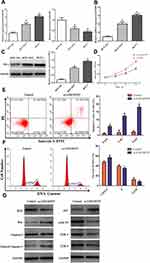
We identified that 605 lncRNAs (442 up-regulated and 163 down-regulated), 3205 mRNAs (2072 up-regulated and 1133 down-regulated) and 104 miRNAs (77 up-regulated and 27 down-regulated) were differentially expressed in BC vs non-tumor tissues (Table S1). The distribution of all DEGs on the two dimensions of -log (FDR) and logFC was depicted in the volcano plot (Figure 1A). For the heatmap shown in Figure 1B, the numerical data represented the expression profile of DEGs.
Construction of the ceRNA Network
We focused on the relationship between DElncRNA, DEmRNA and DEmiRNA to construct the ceRNA network. First, we integrated reliable online databases and prediction website to determine miRNA and target interactions. 16 DEmiRNAs might interact with 9 DElncRNAs. Second, 82 of the 3205 DEmRNAs might have target relationship with 15 DEmiRNAs. Finally, based on the above relationship, we constructed 143 lncRNA-miRNA-mRNA relationships (Figure 2A).
Functional Enrichment Analysis of DEmRNAs in the ceRNA Network
We performed functional enrichment analysis for DEmRNAs through DAVID database. As shown in Figure 2B, abundant biological processes (BP) were mainly related to transcription, DNA-templated, signal transduction and negative regulation of apoptosis. Cell component (CC) mainly enriched in the incaveola, plasma membrane, axon. For molecular function (MF), they mainly enriched in the transcriptional activator activity, core promoter sequence-specific DNA binding and transcriptional factor activity (Table S2). KEGG pathway analysis showed that these DEmRNAs were mainly involved in transcriptional misregulation in cancer, aldosterone-regulated sodium reabsorption, and prostate cancer (Table 2).
 |
Table 2 KEGG Pathway Analysis of the DEmRNAs in the ceRNA Network |
Construction of the PPI Network
To clarify the potential relationship between DEmRNAs in the ceRNA network, PPI network was constructed through STRING database. 82 nodes and 75 edges were shown with an average node degree of 1.83 and a local clustering coefficient of 0.37 (Figure 2C). Cytohubba was also used to select hub-gene, and 12 methods including comprehensive MCC were used for analysis. The ones with higher frequency were identified as hub-gene, indicating that GATA3, PPARG, ESR1 were the TOP3.
Target Prediction and Bioinformatics Analysis of AKBA
A total of 11 potential targets for AKBA were predicted through searching the BATMAN-TCM database, including ESR1, PGR, AR, NR3C1, HMGCR, ITGB2, ANXA1, ITGAL, CYP19A1, HDAC2 and NR3C2, among which ESR1 scored the highest. To further characterize the potential functional pathways of AKBA, we performed KEGG analysis for these targets. 11 targets were mainly enriched in the cell growth and death, endocrine system, natural killer cell mediated cytotoxicity and estrogen signaling pathway. Disease analysis indicated that AKBA had the potential therapeutic effects of brain injury, BC, endocrine independent cancer, estrogen disorders, and so on. In the view of ingredient-target-disease association network (Figure 2D), AKBA might regulate the ESR1, PGR and CYP19A1 in the treatment of BC.
LINC00707 Was Upregulated While miR-206 Was Downregulated in MCF-10AT Cells
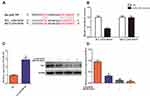
The expression of LINC00707 and miR-206 was measured using qRT-PCR in mammary cell lines including MCF-10AT, MCF-7 and MCF-10A cells. As shown in Figure 3A, compared with MCF-10A cells, LINC00707 expression was significantly upregulated, while miR-206 was downregulated in MCF-10AT and MCF-7 cells (P<0.05), indicating that LINC00707 and miR-206 might be involved in the occurrence and development of breast tumor.
Estrogen Receptor-α mRNA and Protein Were Increased in MCF-10AT Cells
The results of qRT-PCR showed that ESR1 expression was significantly increased in MCF-10AT and MCF-7 cells compared with MCF-10A cells (P<0.05) (Figure 3B). Meanwhile, ER-α protein expression was also increased in MCF-10AT and MCF-7 cells (P<0.05) (Figure 3C).
Knockdown of LINC00707 Inhibited the Proliferation of MCF-10AT Cells
To explore whether LINC00707 can affect MCF-10AT cells proliferation, CCK8 assay was carried out in our study. As shown in Figure 3D, downregulation of LINC00707 significantly inhibited the proliferation of MCF-10AT cells compared with the negative control group (P<0.05). The results demonstrated that downregulation of LINC00707 can inhibit MCF-10AT cell proliferation.
Inhibition of LINC00707 Induced MCF-10AT Cells Apoptosis and Cells Cycle Arrest
Flow cytometry was used to assesse the effect of LINC00707 on MCF-10AT cell apoptosis. The results indicated that inhibition of LINC00707 induced MCF-10AT apoptosis (Figure 3E). In addition, inhibition of LINC00707 increased the ratio of cells in G1 phase whereas decreased those in the S and G2 phase (Figure 3F). The results of Western blotting showed that compared with the control group, the protein expression of Bcl2 was decreased, while Bax and cleaved caspase-3 expression were increased in the transfection si-LINC00707 group. Meanwhile, the protein expression of Cyclin D1, CDK4 and CDK6 was reduced, and P27 was increased in MCF-10AT cells transfected with si-LINC00707 (Figure 3G). These data indicated that inhibition of LINC00707 enhanced MCF-10AT cells apoptosis and repressed cells cycle in G1 phase.
LINC00707 Functions as a ceRNA for miR-206 and Indirectly Modulates the Expression of ER-α
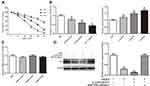
LINC00707 levels were negatively associated with miR-206 expression in MCF-10AT cells. Meanwhile, bioinformatics analysis revealed that miR-206 is a potential target of LINC00707 in breast tumor. The binding sites between LINC00707 and miR-206 are shown in Figure 4A. The dual-luciferase reporter assay indicated that miR-206 mimics can impair the luciferase activity of the WT LINC00707 (P<0.05), but not of the MUT LINC00707 (Figure 4B). In addition, qRT-PCR results showed that the expression of miR-206 increased in MCF-10AT cells transfected with si-LINC00707 compared with transfected with NC, indicating that miR-206 is a target of LINC00707 in MCF-10AT cells (Figure 4C). According to previous research, ESR1 was a direct target of miR-206.19,20 The expression of ER-α protein in MCF-10AT cells was measured (Figure 4D). Compared with NC group, ER-α protein expression was decreased in MCF-10AT cells transfected with si-LINC00707 or miR-206 mimics (P<0.05).
AKBA Inhibited MCF-10AT Cells Growth
CCK8 assay was performed to determine the viability of MCF-10AT cells exposed to the AKBA (5, 10, 20, 30, 40 μM). As shown in Figure 5A, AKBA could significantly inhibit MCF10AT cell growth in dose- and time-dependent manner. The 50% inhibitory concentration (IC50) of AKBA on MCF-10AT cells were about 52.63μM (12h), 22.78μM (24h) and 12.83μM (48h). Therefore, 15μM, 20μM and 25μM AKBA concentration were selected for 24h in subsequent cell experiments.
Effect of AKBA on LINC00707, miR-206 and ESR1 in MCF-10AT Cells
We measured the expression of LINC00707, miR-206 and ESR1 in MCF-10AT cells after AKBA treatment. Compared with the control group, LINC00707 and ESR1 expression were significantly decreased, while miR-206 expression was increased in different dose of AKBA treated groups (P<0.05) (Figure 5B). However, the ESR1 mRNA levels were not altered by AKBA in MCF-10AT cells (Figure 5C).
Effect of AKBA on ER-α Protein in MCF-10AT Cells
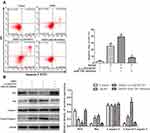
The si-LINC00707 and miR-206 inhibitor were transfected into MCF-10AT cells, respectively, and ER-α protein expression was detected by western blotting. Compared with the control group, ER-α protein expression was decreased in AKBA treated group (P<0.05). However, the ESR1 mRNA levels were not altered by AKBA, suggesting that AKBA may modulate the protein translation of ESR1, possibly through miRNAs. The protein level of ER-α was more significantly reduced in MCF-10AT cells transfected with si-LINC00707. When miR-206 was down-regulated in MCF-10AT cells, ER-α protein expression was increased, and these effects were nearly reversed by treatment with AKBA (Figure 5D). Taken together, AKBA inhibited ER-α protein expression through LINC00707/miR-206 in MCF-10AT cells.
AKBA Promoted MCF-10AT Cells Apoptosis
As shown in Figure 6A, MCF-10AT cell apoptosis rates were 1.28%, 24.05%, 38.28% and 9.29% in control, AKBA, AKBA+si-LINC00707 and AKBA+miR-206 inhibitor groups, respectively. Compared with control group, MCF-10AT apoptosis rates were significantly increased in AKBA and AKBA+si-LINC00707 group, while the miR-206 inhibitor could reverse some of the apoptotic effects of AKBA on MCF-10AT cells. The western-blot results showed that compared with the normal group, the expression of Bcl2 was decreased, while Bax and cleaved caspase-3 protein expression were significantly increased in AKBA and AKBA+si-LINC00707-treated group (Figure 6B). This suggested that AKBA promotes MCF-10AT cells apoptosis.
AKBA Induced Cell Cycle Arrest in MCF-10AT Cells
The flow cytometry results showed that the cell percentages of G0/G1, S and G2/M phases in control group were 45.19%, 40.23% and 14.58%, respectively. Whereas the cell percentages of G0/G1, S and G2/M phases were 61.94%, 31.19% and 6.87% in AKBA group (Figure 7A). Compared with control group, the cell percentages of G0/G1 phase were significantly increased in MCF-10AT cells transfected with si-LINC00707, correspondingly, the cells remaining in S phase and G2/M phase were reduced. However, the biological effect of AKBA inducing G0/G1-phase cell cycle arrest basically disappeared in MCF-10AT cells transfected with miR-206 inhibitors. Furthermore, as shown in Figure 7B, the protein expression of Cyclin D1, CDK4 and CDK6 which are mainly involved in driving the transition from G0/G1 to S phase were markedly reduced in AKBA combination transfection si-LINC00707 MCF-10AT cells. The results suggested that AKBA induces G1-phase cell cycle arrest in MCF-10AT cells.
Discussion
BC is the most common female cancer in the vast majority of countries (140/184), accounting for a quarter of all women diagnosed with cancer. 2018 GLOBOCAN estimated that there were 2088,849 (11.6%) new cases of BC and 626,679 (6.6%) deaths from the disease in this year.21 In recent years, Genome-wide association studies have identified nearly 6500 disease- or trait-predisposing single nucleotide polymorphisms (SNPs), 93% of which are located in non-coding region.22 A CeRNA hypothesis was proposed for the new pattern of gene expression regulation that could be used to further understand the mechanisms of various diseases including cancer. In the present study, we identified 3205 DEmRNAs, 104 DEmiRNAs, and 605 DElncRNAs in BC tissues compared with normal tissues from TCGA, and constructed 143 lncRNA-miRNA-mRNA relationships.
AKBA is a derivative of boswellic acid, which is the main component of a gum resin from Boswellia serrata. AKBA has been used to treat a number of inflammatory diseases, including arthritis, ulcerative colitis, Crohn’s disease, and bronchial asthma.23,24 In addition, AKBA was suggested to prevent cancer progression through the modulation of expression of many proteins and genes involved in apoptosis, angiogenesis and cell growth.20,25 We applied batman-TCM Online database to predict the potential targets, pathways and diseases of AKBA. From the view of ingredient-target-pathway/disease association network, AKBA may regulate the ESR1 in the treatment of BC through the cell growth and death and estrogen signaling pathway.
Most of miRNAs act as gene expression regulators by imperfect or near-perfect base pairing with the 3′-UTR of their target mRNAs, although there may be binding sites for miRNAs at 5ʹ-UTR or extron region of mRNA. After binding, target mRNAs are silenced due to spatial structural alternation.26,27 It is well known that lncRNAs are able to serve as ceRNAs to modulate the expression of genes by sponging miRNAs, thus titrating available miRNAs and contributing to tumorigenesis, including breast carcinogenesis.28,29 In vivo and in vitro studies have demonstrated that LINC00707 may act as an oncogene and competitively bind multiple miRNAs (included miR-206 and miR-876), thus reducing the inhibition of their mRNA targets, and promoting tumor cells proliferation and metastasis.29,30 Several studies have reported that miR-206 was considered to be tumor suppressor, and its expression was frequently down-regulated in various human malignancies. MiR-206 level was significantly downregulated, and overexpression of miR-206 inhibited proliferation, migration and angiogenesis of BC cells through regulating ER-α.31 ER-α plays key roles not only in cell differentiation, motility, invasion, proliferation and cell survival, but also in the advancement and metastasis of solid tumors. The MCF10-AT cell derived from xenograft-passaged MCF10A-ras cells and generated carcinomas in about 25% of xenografts, representing the transition from normal epithelium to malignant cancer.32,33 The qRT-PCR results indicated that LINC00707, miR-206 and ESR1 were dysregulated during the early breast neoplasias and persisted in BC. Our present study showed that down-regulation of LINC00707 was able to suppress cell proliferation, induce apoptosis and cell cycle arrest of MCF-10AT cells, suggesting that LINC00707 may serve as a potential target for BC treatment.
Here, we demonstrated that AKBA can inhibit MCF10AT cell growth in dose-, time-dependent manner. Meanwhile, it could promote cells apoptosis and induce GO/G1 phase cell cycle arrest in MCF-10AT cells. We examined protein level of ER-α, and specifically found that ER-α protein expression was dose-dependently altered by AKBA in MCF-10AT cells, concomitant with the changes in cell proliferation. However, the ESR1 mRNA levels were not altered by AKBA, suggesting that AKBA may modulate the protein translation of ESR1, possibly through miRNAs. And then we found that the expression of LINC00707 was decreased and miR-206 was increased in the AKBA treated groups. Compared with the control group, ER-α protein expression was decreased in AKBA treated group. At the same time, the effect was more significant in MCF-10AT cells transfected with si-LINC00707. When miR-206 was down-regulated in MCF-10AT cells, ER-α protein expression was increased, and these effects were nearly reversed by treatment with AKBA. Taken together, AKBA inhibited ER-α protein expression through LINC00707/miR-206 in MCF-10AT cells (Figure 8). These findings suggested that AKBA may be a promising drug for breast precancerous lesions.
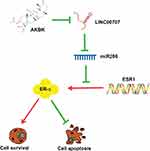 |
Figure 8 Schematic of the model. AKBA inhibits breast precancerous lesions MCF-10AT cells via regulation of LINC00707/miR-206/ER-α signalling. |
Acknowledgments
The authors gratefully acknowledge TCGA for open access to their database. This work was supported by the National Natural Science Foundation of China (nos. 81803979, 81741130, 81673979, 81473688, and 81373314); the Natural Science Foundation of Guangdong Province, China (nos. 2018A030313393 and 2016A030313114); Science and Technology Program of Guangzhou, China (nos. 201803010051, 201707010245, and 201704020117) and the Fourth Batch of TCM Clinical Outstanding Talent Program of China (no. 444258).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jensen HM, Rice JR, Wellings SR. Preneoplastic lesions in the human breast. Science. 1976;191(4224):295–297. doi:10.1126/science.1246614
2. Gucalp A, Traina TA, Eisner JR, et al. Male breast cancer: a disease distinct from female breast cancer. Breast Cancer Res Treat. 2019;173(1):37–48. doi:10.1007/s10549-018-4921-9
3. Maher B. ENCODE: the human encyclopaedia. Nature. 2012;489(7414):46–48. doi:10.1038/489046a
4. Samaeekia R, Adorno-cruz V, Bockhorn J, et al. miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clin Cancer Res. 2017;23(4):1091–1103. doi:10.1158/1078-0432.CCR-16-0943
5. Muhammad N, Bhattacharya S, Steele R, et al. Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget. 2016;7(36):58595. doi:10.18632/oncotarget.11193
6. Leonardo S, Laura P, Yvonne T, Lev K, Pier Paolo P. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
7. Zhou W, Ye XL, Xu J, et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10(483):eaak9557. doi:10.1126/scisignal.aak9557
8. Muhammad N, Steele R, Isbell TS, et al. Bitter melon extract inhibits breast cancer growth in preclinical model by inducing autophagic cell death. Oncotarget. 2017;8(39):66226. doi:10.18632/oncotarget.19887
9. Bhattacharya S, Muhammad N, Steele R, et al. Bitter melon enhances natural killer–mediated toxicity against head and neck cancer cells. Cancer Prev Res. 2017;10(6):337–344. doi:10.1158/1940-6207.CAPR-17-0046
10. Muralidhar Y, Prasad TNK, Kumar TVC, et al. Antibacterial, anti-inflammatory and antioxidant effects of acetyl-11-α-keto-β-boswellic acid mediated silver nanoparticles in experimental murine mastitis. IET Nanobiotechnol. 2017;11(6):682–689. doi:10.1049/iet-nbt.2016.0204
11. Chen M, Wang M, Yang Q, et al. Antioxidant effects of hydroxysafflor yellow A and acetyl-11-keto-β-boswellic acid in combination on isoproterenol-induced myocardial injury in rats. Int J Mol Med. 2016;37(6):1501–1510. doi:10.3892/ijmm.2016.2571
12. Bini Araba A, Ur Rehman N, Al-araimi A, et al. New derivatives of 11-keto-β-boswellic acid (KBA) induce apoptosis in breast and prostate cancers cells. Nat Prod Res. 2019:1–10. doi:10.1080/14786419.2019.1593165
13. Al-harrasi A, Hussain H, Hussain J, et al. Two pyrolysate products from Omani frankincense smoke: first evidence of thermal aromatization of boswellic acids. J Anal Appl Pyrolysis. 2014;110:430–434. doi:10.1016/j.jaap.2014.10.007
14. Li W, Liu J, Fu W, et al. 3-O-acetyl-11-keto-β-boswellic acid exerts anti-tumor effects in glioblastoma by arresting cell cycle at G2/M phase. J Exp Clin Cancer Res. 2018;37(1):132. doi:10.1186/s13046-018-0805-4
15. Csuk R, Barthel-niesen A, Barthel A, et al. 11-keto-boswellic acid derived amides and monodesmosidic saponins induce apoptosis in breast and cervical cancers cells. Eur J Med Chem. 2015;100:98–105. doi:10.1016/j.ejmech.2015.06.003
16. Liu Y, Wang S, Xu Q, et al. Acetyl-11-keto-β-boswellic acid suppresses docetaxel-resistant prostate cancer cells in vitro and in vivo by blocking Akt and Stat3 signaling, thus suppressing chemoresistant stem cell-like properties. Acta Pharmacol Sin. 2019;40(5):689–698. doi:10.1038/s41401-018-0157-9
17. Law CW, Alhamdoosh M, Su S, Smyth GK, Ritchie ME. RNA-seq analysis is easy as 1-2-3 with limma, glimma and edgeR. F1000res. 2016;5:1408. doi:10.12688/f1000research
18. Liu Z, Guo F, Wang Y, et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechanism of traditional Chinese medicine. Sci Rep. 2016;6:21146. doi:10.1038/srep21146
19. Adams BD, Henry F, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21(5):1132–1147. doi:10.1210/me.2007-0022
20. Chen X, Yan Q, Li S, et al. Expression of the tumor suppressor miR-206 is associated with cellular proliferative inhibition and impairs invasion in ERα-positive endometrioid adenocarcinoma. Cancer Lett. 2012;314(1):41–53. doi:10.1016/j.canlet.2011.09.014
21. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
22. Elizabeth P. The biology of genomes. Disease risk links to gene regulation. Science. 2011;332(6033):1031. doi:10.1126/science.332.6033.1031
23. Syrovets T, Büchele B, Krauss C, Laumonnier Y, Simmet T. Acetyl-boswellic acids inhibit lipopolysaccharide-mediated TNF-α induction in monocytes by direct interaction with IκB kinases. J Immunol. 2005;174(1):498–506. doi:10.4049/jimmunol.174.1.498
24. Yasunari T, Haruyo I, Vladimir B, Aggarwal BB. Acetyl-11-keto-beta-boswellic acid potentiates apoptosis, inhibits invasion, and abolishes osteoclastogenesis by suppressing NF-kappa B and NF-kappa B-regulated gene expression. J Immunol. 2006;176(5):3127–3140. doi:10.4049/jimmunol.176.5.3127
25. Xue X, Chen F, Liu A, et al. Reversal of the multidrug resistance of human ileocecal adenocarcinoma cells by acetyl-11-keto-β-boswellic acid via downregulation of P-glycoprotein signals. Biosci Trends. 2016;10(5):392–399. doi:10.5582/bst.2016.01115
26. Gao Q, Zheng J. Ginsenoside Rh2 inhibits prostate cancer cell growth through suppression of micro RNA-4295 that activates CDKN 1A. Cell Prolif. 2018;51(3):e12438. doi:10.1111/cpr.2018.51.issue-3
27. Forman JJ, Legesse-miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets dicer within its coding sequence. Proc Natl Acad Sci USA. 2008;105(39):14879–14884. doi:10.1073/pnas.0803230105
28. Jiao D, Li Z, Zhu M, et al. LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC). Am J Cancer Res. 2018;8(4):748.
29. Li T, Li Y, Sun H. MicroRNA-876 is sponged by long noncoding RNA LINC00707 and directly targets metadherin to inhibit breast cancer malignancy. Cancer Manag Res. 2019;11:5255. doi:10.2147/CMAR.S210845
30. Shao H, Li Q, Shi T, Zhang G, Shao F. LINC00707 promotes cell proliferation and invasion of colorectal cancer via miR-206/FMNL2 axis. Eur Rev Med Pharmacol Sci. 2019;23(9):3749–3759. doi:10.26355/eurrev_201905_17801
31. Naoto K, Tatsuya T, Hiroshi S, Yoshitaka F, Hiroko Y. miR-206 expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68(13):5004. doi:10.1158/0008-5472.CAN-08-0180
32. Pilat MJ, Christman JK, Brooks SC. Characterization of the estrogen receptor transfected MCF10A breast cell line 139B6. Breast Cancer Res Treat. 1996;37(3):253–266. doi:10.1007/BF01806507
33. Shekhar PV, Chen ML, Werdell J, et al. Transcriptional activation of functional endogenous estrogen receptor gene expression in MCF10AT cells: a model for early breast cancer. Int J Oncol. 1998;13(5):907.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.