Back to Journals » OncoTargets and Therapy » Volume 12
Abietic acid suppresses non-small-cell lung cancer cell growth via blocking IKKβ/NF-κB signaling
Authors Liu X , Chen W , Liu Q, Dai J
Received 21 December 2018
Accepted for publication 3 April 2019
Published 20 June 2019 Volume 2019:12 Pages 4825—4837
DOI https://doi.org/10.2147/OTT.S199161
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Xueping Liu,* Wei Chen,* Quanxing Liu, Jigang Dai
Department of Thoracic Surgery, Xinqiao Hospital, Army Medical University (Third Military Medical University), Chongqing 400037, People’s Republic of China
*These authors contributed equally to this work
Background: Abietic acid (AA) is one of the terpenoids, which are multifunctional natural compounds. It has been reported that AA possesses favorable therapeutic effects on inflammation and obesity.
Method: In the present study, we determined the inhibitory effect of AA on the proliferation and growth of non-small-cell lung cancer (NSCLC) cell lines for the first time. Then, flow cytometry and Western blot analysis were applied to determine the cell apoptosis and cell cycle. Finally, surface plasmon resonance, molecular docking and molecular dynamics (MD) simulation were performed to explore the underlying molecular mechanisms.
Results: In vitro experiments indicated that AA displays significant anti-proliferative, cell cycle arresting and pro-apoptotic activities. Mechanistically, AA abrogated tumor necrosis factor-α induced phosphorylation of IκB kinase (IKKα/β) (Ser176/180) and IkBα (Ser32), and inhibited the nuclear translocation of nuclear factor‐κB. Moreover, we found that the activities of AA against NSCLC cells were mediated by its IKKβ inhibition. Molecular docking and MD simulations demonstrated that the mechanism of action between AA and IKKβ was through hydrophobic interactions.
Conclusion: Our data indicate that AA could be a promising lead compound for the discovery of novel IKKβ inhibitors and potential agents for the treatment of NSCLC.
Keywords: abietic acid, hydrophobic interaction, IKKβ inhibitor, NSCLC
Introduction
Lung cancer is the most commonly diagnosed cancer (11.6% of the total cases) and the leading cause of cancer death (18.4% of the total cancer deaths) worldwide.1 Non-small-cell lung cancer (NSCLC) accounts for >85% of all lung cancer cases, for which the predicted 5-year survival rate is extremely poor (15.9%).2 Molecularly targeted therapies, such as EGFR and anaplastic lymphoma kinase (ALK) inhibitors, have brought remarkable improvements to the survival and prognosis for NSCLC patients; however, the overall outcome of current therapies for NSCLC is still unsatisfactory.3 The most frequent oncogene-driven mutations in NSCLC patients are of EGFR (17%) and ALK (7%), but up to 69% of advanced patients may have actionable molecular targets, meaning over 30% cannot benefit from target therapies.4 Besides, the frequency and distribution of EGFR mutations are quite variable across the globe that extremely restrains the clinical benefits of EGFR kinase inhibitors for the treatment of Caucasian NSCLC patients. According to the statistics, the number of NSCLC patients harboring EGFR activation mutations in East Asia was much more than in the United States and Europe.5 The current standard first-line treatment for patients with advanced NSCLC is platinum-based doublet chemotherapy,6 however there is still an urgent need to identify more effective drugs with lower toxicity for the treatment of NSCLC.
Natural products (NPs) have proven a reliable and consistent source for medicinal chemistry research and drug discovery in recent decades.7 NPs display a broad-spectrum of biological effects, including well-studied properties such as anti-inflammatory, anti-microbial, and especially anti-tumor activities. Approximately 60% of clinically approved antitumor drugs have reportedly originated from NPs.8 Paclitaxel, vinblastine and doxorubicin, for example, are the most well-known chemotherapies in clinical use at present.9 Compared to alkylating antineoplastic agents or antimetabolite antitumor agents, NP-derived antitumor drugs offer a remarkable advantage of having lower toxicity to normal tissues. On the other hand, NPs are often perceived as chemically complex, differing from synthetic drug-like molecules in many respects. Fragment-like NPs with innovative scaffolds have great promise for their use as starting points in chemical biology and medicinal chemistry fields.
In recent years, it is well recognized that IκB kinase (IKK)/nuclear factor-κB (NF-κB) signal pathway plays a crucial role in the pathogenesis of a number of human diseases, including asthma, neurodegeneration, inflammation and cancers. In the canonical NF-κB pathway, cell is stimulated by various stimuli such as tumor necrosis factor-α (TNF-α), IL-1 and lipopolysaccharides. Subsequently, these activators trigger IKKβ phosphorylated by TGF-β-activated kinase 1, then the phosphorylation signals were transferred to the inhibitory IκB proteins, leading to their rapid ubiquitination and proteasome-mediated degradation, which finally makes NF-κB nuclear translocated and various NF-κB-related tumor-promoting genes transcribed including Bcl-2, Cyclin D1, survivin, and so on. Based on early studies that identified IKKβ as a key tumor promoter via driving the classical NF-κB signaling activation, several potent synthetic inhibitors of IKKβ have developed. However, the reports on the NP-derived IKKβ inhibitors are still pretty rare.10
Abietic acid (AA) is an abietane diterpenoid compound mainly derived from Pimenta racemosa var. grissea, which reportedly possesses anti-allergenic, anti-inflammatory, anti-obesity and anti-convulsant activities.11–14 In recent studies, researchers found AA and its analogs display novel anti-tumor effects as potential adjuvant therapy agents.15–17 Hsieh reported that AA not only effectively suppressed melanoma cancer cell metastasis in both in vitro and in vivo models, it also improved the efficacy of taxol against B16F10 cells.15 Furthermore, Yoshida reported that AA inhibits MRP2- or BCRP-mediated membrane transport and their interaction with substrate.18 However, beyond these, evaluations of the anti-proliferative activity of AA have been extremely rare, the complete anti-cancer potential of AA has yet to be revealed.
In our study, we first screened the killing ability of AA on six NSCLC cell lines. Cell proliferation (MTS assay) and clone formation results suggested AA remarkably inhibited the proliferation and growth of NSCLC cells. Flow cytometry and Western blot analysis data indicated that AA effectively arrested the cell cycle of NSCLC cells at the G0/G1phase and induced apoptosis. Furthermore, mechanistic investigation results revealed AA was a potential IKKβ inhibitor. IKKβ overexpression by specific plasmid was conducted. The phenotype results showed the anti-cancer role of AA was greatly impaired, further indicating the activity of AA is mediated through the IKKβ/NF-κB signaling inhibition. At last, the interaction between IKKβ and AA was simulated through molecular docking and molecular simulations, which demonstrated the hydrophobic interaction may contribute to their binding interaction.
Materials and methods
Cell culture and reagents
The human pulmonary epithelial cells Beas-2B and NSCLC cell lines A549, H460, HCC827, H1650, PC-9 and H1975 were obtained from Shanghai Institute of Biosciences and Cell Resources Center (Chinese Academy of Sciences, Shanghai, China). The cells were routinely cultured in RPMI-1640 or DMEM (Gibco/BRL lifeTechnologies, Eggenstein, Germany) supplemented with 10% fetal bovine serum FBS (Hyclone, Logan, UT, USA), 1% penicillin/streptomycin solution in a humidified atmosphere of 5% CO2 at 37°C. The primary antibodies used in this study, including p-IKKβ (Ser176/180), IKKβ, p-IκBα (Ser32), IKBα, Bcl-2, cleaved-PARP, Cyclin D1, Cdk-4 and Actin were all purchased from Cell Signaling Technology (Danvers, MA, USA). Goat anti-rabbit IgG-horseradish peroxidase (HRP) secondary antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA); AA was purchased from Solorbio (Beijing, China). and the purity of AA detected by HPLC was over 99% (Figure S1). Human recombinant IKKα and IKKβ protein was purchased from Abcam (Cambridge, UK), and their purities were both >85% assessed by SDS-PAGE (Manufacturer datasheet). The pcDNA-Ikkb-FLAG WT plasmid was purchased from Addgene (MA, USA).
 | Figure S1 The purity of AA detected by HPLC. |
Transfection of plasmid
For plasmid transfections, cells were seeded overnight and plasmids were transfected with X-treme GENE HP DNA Transfection Reagent (Roche Applied Science, Shanghai, China) according to the manufacturer’s instructions. Cells were harvested for protein extraction after 48 hrs of transfection.
Cell viability assay
The cell viability was measured by the Cell Titer 96 Aqueous Non-Radioactive Cell Proliferation Assay Kit (Promega, Fitchburg, WI, USA). 5×103 cells were seeded in 96-well plates overnight, then the culture medium was removed and all cells were treated with different concentrations of chemicals for 72 hrs. The absorbance value of each well was determined by a microplate reader at 490 nm. Each experiment was repeated for three times.
Clonogenic assay
PC-9 and H1975 cells (1,000 cells/well) were seeded in 6-well cell culture plate for 24 hrs, then DMSO and AA (20 or 40 µM) were added to the culture medium. After 24 hrs, the culture medium was replaced with fresh culture medium for 14 days. Colonies were removed the medium, washed with PBS twice, fixed with methanol for 15 min, then washed with PBS three times, and finally stained with crystal violet for 15 mins. Colonies containing >50 cells were counted, and visualized colonies were then photographed.
Cell apoptosis analysis
Cell apoptosis was detected by flow cytometer analysis and Western blotting. For flow cytometer analysis of apoptosis, cells treated as indicated for 24 hrs were harvested by trypsin, washed twice by PBS, and re-suspended in 100 μL 1× binding buffer. 5 μL FITC Annexin V and propidium iodide (PI) (556547, BD Biosciences, Franklin Lakes, NJ, USA) were added to the cell suspension and then incubated for 15 mins at room temperature. After dilution with 400 μL binding buffer, the samples were analyzed by ACS Calibur flow cytometer (BD Biosciences). For apoptosis by Western blotting, cleaved caspase 3, cleavage of PARP (poly ADP-ribose polymerase), Bcl-2 and BCl-xL were analyzed.
Cell cycle analysis
Cells (3×105 cells/well) were seeded in 6-well plates and allowed to adhere overnight. The next day, the cells were treated with different compounds as indicated for 24 hrs. Then, the cells were trypsinized, washed, and fixed in 75% ice-cold ethanol at 4°C overnight. After centrifugation, the pellets were washed with cold PBS, suspended in 500 μL PBS with 50 mg/mL PI and incubated at 4°C for 30 mins in the dark. Then, cell suspension was subjected to a FACS Calibur instrument (Becton Dickinson FACSCalibor; BD Biosciences).
Western blotting
After treated as indicated for 24 hrs, the cells were washed once by PBS. The cell lysates were quantitated by BCA protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA). Equal amounts of proteins were separated by SDS-PAGE and transferred to PVDF membrane. After being blocked with 5% non-fat dry milk in TBST for 1.5 hrs, membranes were incubated with a 1:1,000 dilution of specific primary antibody overnight at 4°C and the secondary antibody conjugated with HRP (1:5,000; Santa Cruz Biotechnology) for 2 hrs, the immunoreactive bands were visualized with enhanced chemiluminescence (EMD Millipore, Billerica, MA, USA) in Amersham Imager 600 system (GE Healthcare Life Sciences, Shanghai, China).
Surface plasmon resonance (SPR) analysis
SPR experiments were performed on a ProteOn XPR36 Protein Interaction Array system (Bio-Rad Laboratories). Briefly, IKKβ solution in PBST (5 mM, pH 7.4) at a concentration of 1 mg/mL was diluted to 30 μg/mL with sodium acetate buffer (pH 4.5). The chip was activated with EDC/NHS (10 μL/min for 600 s). Then, IKKβ was loaded (5 μL/min for 400 s) and immobilized covalently. Approximately 8,000 RU of IKKβ was immobilized on the chip. Any excess of unbound IKKβ was removed by flowing PBS solution (5 mM, pH 7.4, with 5%, w/v, DMSO). AA was prepared as 20–100 μM solution in PBS solution (5 mM, pH 7.4, with 5%, w/v, DMSO), and injected (10 μL/min for 100 s). Five concentrations were injected simultaneously at a flow rate of 30 μM/min for 120 s of association phase, followed with 120 s of dissociation phase at 25°C. The final graph was obtained by subtracting blank sensorgrams from the duplex or quadruplex sensorgrams. Data were analyzed by ProteOn manager software.
Construction of the initial structure of IKKβ/AA complex
The crystal structure of IKKβ was retrieved from the Protein Data Bank (PDB) database (PDB code: 4KIK).19 The crystal structure of IKKβ was refined by UCSF Chimera program,20 including removing all water molecules, non-bonded hetero-atoms, and adding missing hydrogen atoms. Then, the binding pose of AA in the allosteric pocket between the kinase domain and ubiquitin-like domain was predicted by the AutoDock program as previous study.21,22 The program and AA were processed by AutoDockTools 1.5.6 program.21 A grid box size of 22.5 Å ×22.5 Å ×22.5 Å dimensions, with a spacing of 0.375 Å between the grid points, was selected and covered the entire allosteric pocket. Lamarckian Genetic Algorithm was applied to conformational sampling with trials of 200 dockings, maximum number of evaluation 25,000,000 and other settings were set as default. The lowest binding energy conformation was used for molecular dynamics (MD) simulation analysis.
Molecular dynamics (MD) simulation
The IKKβ/AA complex provided by molecular docking was used as the initial structure for the MD simulations. The partial atomic charges for AA were evaluated by the restrained electrostatic potential (RESP) method based on HF/6-13G* basis set. Then, the complex was processed by the LEaP module in AmberTools14. The ff14SB force field was assigned to the IKKβ residues, and General Amber Force Field 2 was used to describe the AA.23,24 Thereafter, the complex was immersed in a rectangular box of water molecules (TIP3P). Finally, counterions were added to neutralize the modeled system.
Before the productive MD simulations, the system was minimized by 8,000 steps of steepest descent followed by 8,000 steps of conjugate gradient to remove the bad contacts in the modeled structure. Then, the system was heated up from 0 to 300 K in 300 ps at a constant force of 6.0 kcal/mol Å−2 to constrain the protein atoms. After heating, 200 ps density procedure at 300 K and 200 ps equilibration in the isothermal-isobaric (NPT) ensemble were applied to raise the density and control the temperature of the system. In the end, a 100 ns MD simulation was performed at a constant temperature of 300 K and a constant pressure of 1 atm. Coordinates were recorded every 4 ps and numerical analysis was calculated by CPPTRAJ module in Amber14.25 A total of 500 snapshots extracted from the last 20 ns MD trajectory were applied for the binding free energy calculations by using molecular mechanics/generalized Born surface area (MM/GBSA) method.26
Statistical analysis
The results are presented as the mean ± standard error (SEMs). The statistics were performed using one-way ANOVA in GraphPad Pro (GraphPad, San Diego, CA, USA). P-values <0.05 were considered statistically significant. All the experiments were repeated a minimum of three times. All of the aforementioned experiments were repeated thrice.
Results
AA inhibits NSCLC cell proliferation and colony formation
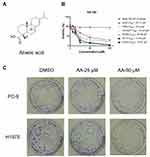
Firstly, the anti-proliferative effects of AA (Figure 1A) against normal human pulmonary epithelial cells (Beas-2B) and six NSCLC cell lines was tested by means of colorimetric MTS assays. As shown in Figure 1B, AA exhibited the most potent inhibitory effect on PC-9 and H1975 cells, with IC50 values of 14.54 μM and 19.97 μM, respectively. Therefore, these two cell lines were chosen for the next experiments. In addition, the status of EGFR mutations of the NSCLC cell lines is listed in Table 1. Combined the EGFR mutational status of NSCLC cell lines with the MTS assay results, it indicated that AA exerted the anti-proliferative effects through an EGFR mutation independent manner.
 | Table 1 EGFR mutational status of the NSCLC cell lines used in the study. This data is according to cancer cell line encyclopedia database |
In addition, colony-forming assays were carried out to evaluate the long-term suppressive effects of AA treatment. After incubating with AA for 2 weeks, cell colonies were stained with crystal violet and counted. Results indicated that AA produced a strong inhibitory effect on the growth of NSCLC cells (Figure 1C).
AA induces NSCLC cell cycle arrest
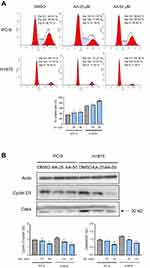
To examine whether AA impacts NSCLC cell cycle distribution, the proportion of cells in G0/G1, S, and G2/M phases was counted by flow cytometry. The results indicated that AA effectively arrested PC-9 and H1975 cells at the G0/G1 phase (Figure 2A). On the other hand, Western blot results suggested that AA dose-dependently down-regulated the expression of G1/S transition regulatory proteins (cyclin D1 and cdk4, Figure 2B). These results suggested that AA effectively arrested the cells in the G0/G1 phase through reducing the expression of cell cycle-related proteins.
AA treatment induces NSCLC cell apoptosis
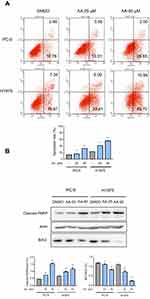
Double-staining with annexin V-FITC and PI was performed to determine whether AA could exert the pro-apoptotic effect on NSCLC cells. Flow cytometry results indicated that compared with DMSO control group, early- and late-stage apoptosis rates of PC-9 and H1975 cells were both greatly increased after treatment with AA for 24 hrs (Figure 3A).
To elucidate the mechanisms of AA-induced apoptosis, we examined the expression levels of the apoptosis-related proteins cleaved caspase-PARP and Bcl-2 by Western blot analysis. Figure 3B indicates the expression of pro-apoptotic molecules cleaved-PARP was significantly upregulated, while the anti-apoptotic protein bcl-2 was downregulated.
AA binds directly to IKKβ and suppresses the IKKβ/Nf-κB signaling pathway in NSCLC cells
The transcription factor NF-κB and its associated regulatory factors, the IKKs, are involved in the pathogenesis of a variety of hematologic and solid tumor malignancies (eg, NSCLC, gastric cancer, breast tumors, etc.). Previously, Thummuri et al showed that AA suppressed activation of the RANKL-induced IKKβ-IκB-NF-κB signaling pathway in a dose-dependent manner.28 Therefore, we speculated that the anti-lung cancer activity of AA may also be involved with inhibition of the IKKβ/NF-κB signaling pathway.
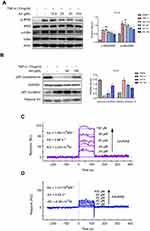
TNF-α, a well-known NF-κB activator, was used to activate the IKKβ/NF-κB pathway. The phosphorylation and total protein expression levels of IKKβ and IκB were analyzed by Western blot. The results showed that AA dose-dependently decreased IKKβ and IκB phosphorylation levels following stimulation with TNF-α (10 ng/mL) for 0.5 and 1 hr, respectively (Figure 4A). As a transcription factor, NF-κB must translocate to the nucleus to regulate target gene transcription. Thus, we also measured the subcellular localization of NF-κB p65 after treatment with AA. The levels of NF-κB p65 subunit in the nuclear and cytosolic fractions were quantified by Western blot. As shown in Figure 4B, AA effectively blocked the nuclear translocation of NF-κB.
To investigate whether AA bound directly to IKKβ, SPR experiments were performed. As shown in Figure 4C, the SPR response value increased gradually with elevated AA concentrations and a low equilibrium dissociation constant (KD) of 32 μM was determined. Besides, to validate the specificity of binding capacity of AA to IKKβ, as well as rule out the artificial effects in our SPR assay, the interaction between AA and IKK α was also determined. The results (Figure 4D) demonstrated that the binding of AA to IKKα was much lower than IKK beta, suggesting AA might be a potential selective IKKβ inhibitor. Collectively, these results indicate that AA bound directly to IKKβ, preventing the phosphorylation of IKKβ and IκB, and the subsequent nuclear translocation of NF-κB.
To clarify the causal relationship between the functional effects and the signaling pathway, IKKβ expressing plasmid was transfected into PC-9 cells (PC-9/IKKβ) and the levels of p-IKKβ and total IKKβ expression were assessed. The results (Figure 5A) showed that IKKβ overexpression successfully increased the protein levels as well as the phosphorylation degree of IKKβ in cells. Consistently, using this overexpressed cell line, we found that overexpression of IKKβ severely impaired AA-induced anti-proliferative and apoptosis effects in PC-9 cells (Figure 5B and C). These findings supported the role of IKKβ in AA-mediated cytotoxicity in NSCLC cells.
Analysis of the AA binding site by molecular modeling
Firstly, molecular docking was used to predict the possible interactions between IKKβ and AA. Next, 100 ns MD simulations were performed to analyze their dynamic behavior. The root-mean square deviations (RMSDs) of all the backbone atoms of IKKβ, and the heavy atoms of AA were analyzed to validate the stability of the IKKβ/AA complex. As shown in Figure 6A, the RMSD values of backbone atoms of IKKβ have a small fluctuation after 25 ns and AA exhibited relative stability during the whole simulation. These findings indicated that through MD simulations we had determined a relatively reasonable binding conformation for our IKKβ/AA complex.
An MD simulation trajectory of the last 20 ns was performed to determine the role of individual residues in protein–ligand recognition patterns. The binding free energies were decomposed into the contributions of each residue based on the MM/GBSA method. As shown in Figure 6B, the 10 highest contributing residues were Ile-371, Lys-310, Leu-376, His-380, Leu-123, Leu-311, Asn-308, Met-384, Val-312 and Leu-307 (Figure 6B). Structural analysis suggested that AA binds to the allosteric pocket of IKKβ to block its activation. The interactions were primarily hydrophobic (Figure 6C). Overall, combining the SPR results with the two simulation methods, we concluded that IKKβ may be the target of AA.
Discussion and conclusions
Despite the tremendous efforts and progress in the past decades, anti-cancer drug development has been considerably hampered by the limited source of chemical scaffolds. NPs consist of a rich source of compounds with abundant structural diversity, and have been extensively explored in the field of anti-cancer drug discovery.29 As the largest class of NPs, terpenoids include approximately 25,000 chemical structures, and a growing number of terpenoids have exhibited anti-cancer activities.30 Celastrol, triptolide and andrographolide are representative terpenoids with anti-cancer activity, and their potential molecular mechanisms are all involved to IKKβ/NF-κB signal pathway.
AA was another promising natural-derived diterpenoids compound, which had been proved to possess significant anti-inflammatory and anti-obesity effect. Oral preparations of AA have been used in clinical studies to treat psoriasis in China, which support the basic safety features of AA in developing it as an anti-tumor agent. In a recent research, Hsieh et al showed that AA could exert inhibitory effects on the metastasis of melanoma cancer.15 However, the anti-lung cancer effects of AA have yet to be investigated. In this study, the inhibitory effects AA against NSCLC cells were evaluated for the first time. Based on our results, AA significantly reduced the proliferation and growth of NSCLC cells, but showed no cytotoxicity against human pulmonary epithelial (Beas-2B) cells. Meanwhile, AA dose-dependently induced cell cycle arrest and apoptosis in NSCLC cells. Given these effects, we believe AA is an effective and low-toxic anti-NSCLC compound.
As a crucial regulator of survival, apoptosis and migration of cancer cells; the IKKβ/NF-κB signaling pathway has been linked to the oncogenic potential of various cancers.31,32 Three potent IKKβ inhibitors, TPCA-1, BMS345541 and ML-120B, were previously evaluated for their anti-cancer effects in preclinical research.33 The chronic toxicity of ML-120B and TPCA-1 found in experiments, however, greatly restricts their further clinical application. In a previous study, Thummuri et al reported that AA alleviated RANKL-induced osteoclastogenesis and inflammation-associated osteolysis through suppressing NF-κB and MAPK signaling.28 In our study, Western blot results suggested AA could also inhibit activation of the IKKβ/NF-κB pathway in NSCLC cells. SPR analysis further confirmed that AA is a direct and potential selective IKKβ inhibitor. Moreover, overexpression of IKKβ rescued the inhibitory effects of AA, implicating the functional effects of AA was highly IKKβ/NF-κB inhibition dependent (Figure 7). Molecular docking and simulation analyses showed that IKKβ inhibition by AA mainly depends on hydrophobic interactions with the hydrophobic cavity of IKKβ. We noticed this result was highly consistent with the extremely hydrophobic structural characteristics of AA, which suggested that to enhance IKKβ inhibitory activity, a modification introducing more hydrogen donors should be performed. Furthermore, introducing a long-chain hydrophilic moiety in the aromatic ring oriented away from the allosteric pocket may effectively increase water solubility and stabilize the binding conformation between AA and IKKβ. This could greatly improve the current poor druggability of AA.
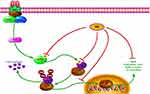 | Figure 7 Schematic illustration of the underlying mechanism of AA’s anti-cancer activity. |
In summary, our results indicate AA significantly inhibits proliferation, arrests the cell cycle and promotes apoptosis of NSCLC cells. Mechanistically, our results demonstrate that AA could directly and selectively bind to IKKβ, preventing the phosphorylation of IKKβ and of the downstream molecule IκB, and reduces the nuclear translocation of NF-κB p65. Moreover, the IKKβ overexpression experiment suggests the anti-NSCLC effects of AA may be mainly mediated by its IKKβ inhibition. Finally, molecular docking and molecular simulations of the AA/IKKβ complex revealed that stable binding between these two molecules depended on hydrophobic interactions. Our results suggest that AA could be a lead compound for the discovery of novel IKKβ inhibitors, as well as a promising drug candidate for the treatment of NSCLC.
Acknowledgments
The study was supported by funding from the General Program of National Natural Science Foundation of China (81472188). We thank The Board institute for publishing these useful and open-access information including the database of cancer cell lines genomic profiles and mutations. We thank the significant efforts of numerous researchers who carried out the original analysis and collection of the data, and we declare they bear no responsibility for the further analysis or interpretation of the data used in our work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546. doi:10.1038/nrc3775
3. Tan WL, Jain A, Takano A, et al. Novel therapeutic targets on the horizon for lung cancer. Lancet Oncol. 2016;17(8):e347–e362. doi:10.1016/S1470-2045(16)30123-1
4. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi:10.1016/S0140-6736(16)30958-8
5. McIntyre A, Ganti AK. Lung cancer – a global perspective. J Surg Oncol. 2017;115(5):550–554. doi:10.1002/jso.24532
6. Rossi A, Di Maio M. Platinum-based chemotherapy in advanced non-small-cell lung cancer: optimal number of treatment cycles. Expert Rev Anticancer Ther. 2016;16(6):653–660. doi:10.1586/14737140.2016.1170596
7. Rodrigues T, Reker D, Schneider P, Schneider G. Counting on natural products for drug design. Nat Chem. 2016;8(6):531–541. doi:10.1038/nchem.2479
8. Sanders K, Moran Z, Shi Z, Paul R, Greenlee H. Natural products for cancer prevention: clinical update 2016. Semin Oncol Nurs. 2016;32(3):215–240. doi:10.1016/j.soncn.2016.06.001
9. Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79(3):629–661. doi:10.1021/acs.jnatprod.5b01055
10. Gamble C, McIntosh K, Scott R, Ho KH, Plevin R, Paul A. Inhibitory kappa B Kinases as targets for pharmacological regulation. Br J Pharmacol. 2012;165(4):802–819. doi:10.1111/j.1476-5381.2011.01608.x
11. Gao Y, Zhaoyu L, Xiangming F, et al. Abietic acid attenuates allergic airway inflammation in a mouse allergic asthma model. Int Immunopharmacol. 2016;38:261–266. doi:10.1016/j.intimp.2016.05.029
12. Kang S, Zhang J, Yuan Y. Abietic acid attenuates IL-1beta-induced inflammation in human osteoarthritis chondrocytes. Int Immunopharmacol. 2018;64:110–115. doi:10.1016/j.intimp.2018.07.014
13. Hwang KH, Ahn JY, Kim S, Park JH, Ha TY. Abietic acid has an anti-obesity effect in mice fed a high-fat diet. J Med Food. 2011;14(9):1052–1056. doi:10.1089/jmf.2010.1471
14. Talevi A, Cravero MS, Castro EA, Bruno-Blanch LE. Discovery of anticonvulsant activity of abietic acid through application of linear discriminant analysis. Bioorg Med Chem Lett. 2007;17(6):1684–1690. doi:10.1016/j.bmcl.2006.12.098
15. Hsieh YS, Yang SF, Hsieh YH, et al. The inhibitory effect of abietic acid on melanoma cancer metastasis and invasiveness in vitro and in vivo. Am J Chin Med. 2015;43(8):1697–1714. doi:10.1142/S0192415X15500962
16. Yoshida N, Takada T, Yamamura Y, Adachi I, Suzuki H, Kawakami J. Inhibitory effects of terpenoids on multidrug resistance-associated protein 2- and breast cancer resistance protein-mediated transport. Drug Metab Dispos. 2008;36(7):1206–1211. doi:10.1124/dmd.107.019513
17. Xu H, Liu L, Fan X, Zhang G, Li Y, Jiang B. Identification of a diverse synthetic abietane diterpenoid library for anticancer activity. Bioorg Med Chem Lett. 2017;27(3):505–510. doi:10.1016/j.bmcl.2016.12.032
18. Gonzalez MA, Correa-Royero J, Agudelo L, Mesa A, Betancur-Galvis L. Synthesis and biological evaluation of abietic acid derivatives. Eur J Med Chem. 2009;44(6):2468–2472. doi:10.1016/j.ejmech.2009.01.014
19. Liu S, Misquitta YR, Olland A, et al. Crystal structure of a human IkappaB kinase beta asymmetric dimer. J Biol Chem. 2013;288(31):22758–22767. doi:10.1074/jbc.M113.482596
20. Pettersen EF, Goddard TD, Huang CC, et al. UCSF chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi:10.1002/jcc.20084
21. Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi:10.1002/jcc.21256
22. Liu H, Liang H, Meng H, Deng X, Zhang X, Lai L. A novel allosteric inhibitor that prevents IKKbeta activation. Medchemcomm. 2018;9(2):239–243. doi:10.1039/c7md00599g
23. Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–3713. doi:10.1021/acs.jctc.5b00255
24. Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi:10.1002/jcc.20035
25. Roe DR, Cheatham TE
26. Sun H, Li Y, Tian S, Xu L, Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys Chem Chem Phys. 2014;16(31):16719–16729. doi:10.1039/c4cp01388c
27.
28. Thummuri D, Guntuku L, Challa VS, Ramavat RN, Naidu VGM. Abietic acid attenuates RANKL induced osteoclastogenesis and inflammation associated osteolysis by inhibiting the NF-KB and MAPK signaling. J Cell Physiol. 2018;234(1):443–453.
29. DeCorte BL. Underexplored opportunities for natural products in drug discovery. J Med Chem. 2016;59(20):9295–9304. doi:10.1021/acs.jmedchem.6b00473
30. Huang M, Lu JJ, Huang MQ, Bao JL, Chen XP, Wang YT. Terpenoids: natural products for cancer therapy. Expert Opin Investig Drugs. 2012;21(12):1801–1818. doi:10.1517/13543784.2012.727395
31. Perkins ND. The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer. 2012;12(2):121–132. doi:10.1038/nrc3204
32. Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi:10.1186/1476-4598-12-86
33. Huang JJ, Chu HX, Jiang ZY, Zhang XJ, Sun HP, You QD. Recent advances in the structure-based and ligand-based design of IKKbeta inhibitors as anti-inflammation and anti-cancer agents. Curr Med Chem. 2014;21(34):3893–3917.
Supplementary material
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.