Back to Journals » OncoTargets and Therapy » Volume 15
Aberrant miR-362-3p is Associated with EBV-Infection and Prognosis in Nasopharyngeal Carcinoma and Involved in Tumor Progression by Targeting JMJD2A
Received 16 June 2021
Accepted for publication 2 September 2021
Published 28 January 2022 Volume 2022:15 Pages 121—131
DOI https://doi.org/10.2147/OTT.S325100
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Xiangyun Wang,* Ping Chen*
Department of Otorhinolaryngology, Dongying People’s Hospital, Dongying, Shandong, 257091, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiangyun Wang
Department of Otorhinolaryngology, Dongying People’s Hospital, No. 317 Nanyi Road, Dongying, Shandong, 257091, People’s Republic of China
Tel/Fax + 86-0546-8901191, Email [email protected]
Background: Many microRNAs (miRNAs) are involved in the progression of nasopharyngeal carcinoma (NPC). This study aimed to examine the expression and clinical significance of microRNA (miR)-362-3p in NPC, especially in Epstein–Barr virus (EBV)-positive patients, and explore its potential mechanism in NPC progression.
Methods: miR-362-3p levels and Jumonji C domain 2A (JMJD2A) mRNA levels were detected by quantitative real-time PCR. The diagnostic value of miR-362-3p to distinguish NPC patients and EBV-positive cases was evaluated using receiver operating characteristic analysis. The association of miR-362-3p with NPC survival was assessed by Kaplan–Meier curves and Cox regression analysis. NPC cell proliferation, migration and invasion were determined using Cell Counting Kit-8 and Transwell assays, respectively. A luciferase reporter assay was used to confirm the interaction between miR-362-3p and JMJD2A.
Results: miR-362-3p expression was decreased in the serum and tissues of NPC patients and had diagnostic value for screening NPC. According to the survival follow-up, NPC survivors had significantly higher miR-362-3p, and miR-326-3p was demonstrated as an independent prognostic indicator of NPC. Interestingly, it is found that EBV-positive NPC patients and cells had significantly lower miR-362-3p compared with EBV-negative NPC patients and cells and had certain ability to distinguish EBV-positive patients. Moreover, miR-362-3p inhibited the proliferation, migration and invasion of both EBV-positive and -negative NPC cells, and these effects might be mediated by targeting JMJD2A.
Conclusion: Abnormal miR-362-3p expression is related to EBV-infection and prognosis in NPC patients and may be involved in NPC progression by targeting JMJD2A.
Keywords: microRNAs, Epstein–Barr virus, prognosis, nasopharyngeal carcinoma, tumor progression, diagnosis
Introduction
Nasopharyngeal carcinoma (NPC) is one of the major squamous cell carcinoma among head and neck squamous cell carcinoma (HNSC), originating from the epithelium of nasopharynx.1 Its incidence and mortality in our country are at a high level worldwide, and it is the most common type of head and neck malignant tumors.2,3 Currently, the preferred treatment for NPC remains radiotherapy, and concurrent chemoradiotherapy has been shown to improve NPC survival.4 The pathogenesis of NPC is unknown and is generally believed to be related to genetic factors, Epstein–Barr virus (EBV) infection, and environmental factors.5,6 EBV is a γ herpesvirus, and it has been demonstrated that EBV infection is one of the major etiological factors of NPC tumorigenesis.7 Therefore, screening populations at high risk of NPC in EBV infected patients is helpful in reducing the occurrence and development of NPC.
microRNAs (miRNAs) are a group of non-coding RNAs 18–25 nucleotides in length that can regulate gene expression.8 There have been numerous studies reporting the important roles of miRNAs in human malignancies.9,10 In addition, microRNA (miR)-362-3p has been found to function as a tumor suppressor in multiple tumor diseases, such as ovarian cancer,11 epithelial ovarian cancer12 and cervical cancer.13 However, its role in NPC remains unknown. In this study, we analyzed the expression levels of miR-362-3p in TCGA and Genomic Data Commons Data Portal databases, and found that it was significantly decreased in HNSC. Therefore, it is necessary to analyze the expression levels of miR-362-3p in clinical NPC patient samples to clarify its role in NPC.
Jumonji C domain 2A (JMJD2A), also named lysine-specific demethylase 4A (KDM4A), has been previously reported as an oncogene that promotes NPC disease progression.14 The present study predicted that JMJD2A has complementary sequences with miR-362-3p, thereby we speculated that there might be a binding between the two. However, whether miR-362-3p is associated with JMJD2A in NPC and whether JMJD2A is able to mediate some kind of biological function that miR-362-3p exerts in NPC are all unknown.
Thus, the purpose of the present study was to measure the expression levels of miR-362-3p in NPC patients, explore the relationship between miR-362-3p and EBV-infection, investigate the clinical value of miR-362-3p in NPC patients, and explore the potential mechanism of miR-362-3p in NPC. This study may provide a novel biomarker for NPC patients and a potential target for NPC therapy.
Methods and Materials
Patient Recruitment and Sample Collection
Serum and tissue samples were collected from 112 NPC patients who were admitted to Dongying People’s Hospital between 2012 and 2016. The inclusion criteria for patients were as follows: 1) patients diagnosed with NPC; and 2) all patients had not received operation, radiotherapy, chemotherapy or antibiotic therapy before biopsy and blood sampling. Patients were excluded if they 1) had psychiatric disorders; 2) had other comorbid tumors; or 3) were not willing to cooperate with treatment. Whole blood was drawn from all patients, and serum was separated by centrifugation at low temperature. Then, a portion of the serum was analyzed for EBV virus capsid antibody (VCA)-IgA, IgG and EBV-DNA, which were used to judge the EBV infection status of the patients; the remaining serum samples were stored at −80°C for RNA extraction. In addition, the serum of 66 healthy controls with no history of malignancy and negative for EBV were collected as controls. Tumor tissue samples and adjacent normal tissues were collected from NPC patients and immediately stored in liquid nitrogen. The clinical characteristics of the patients, including age, gender, EBV infection, smoking, differentiation and TNM stage, were recorded and shown in Table 1. Patients participated in the 5-year survival follow-up, and patient survival was recorded by telephone or face-to-face calls at monthly intervals for subsequent survival analysis. This study was approved by the Ethics Committee of Dongying People’s Hospital (Approval No. #0011831), and written informed consent was obtained from all the participants. This study was conducted in accordance with the Declaration of Helsinki.
 |
Table 1 Association of miR-362-3p with the Clinicopathological Characteristics of NPC Patients |
Cell Culture and Transfection
EBV-negative normal nasopharyngeal epithelial cell line NP69, EBV-negative (EBV-) NPC cell line SUNE1 and EBV-positive (EBV+) NPC cell line C666-1 were obtained from the Cell Bank of Chinese Academy of Science (Shanghai, China). They were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen, USA), supplemented with 10% fetal bovine serum (FBS, Invitrogen). These cell lines were maintained at 37°C in a humidified atmosphere containing 5% CO2.
The miR-362-3p mimic (5’-AACACACCUAUUCAAGGAUUCA-3’), miR-362-3p inhibitor (5’-UGAAUCCUUGAAUAGGUGUGUU-3’), mimic negative control (NC) (5’-UUCUCCGAACGUGUCACGU-3’) and inhibitor NC (5’-CAGUACUUUUGUGUAGUACAA-3’) were transfected into SUNE1 and C666-1 cell lines using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. After 48 h of transfection, the transfected cells were used for subsequent experimental analyses.
RNA Extraction and Quantitative Real-Time PCR
Total RNA from serum, tissues and cells were extracted using Total RNA Purification Kit (Norgen Biotek, Thorold, ON, Canada). Then, 1 μg of total RNA was reverse transcribed to cDNA using the iScript cDNA Synthesis Kit (BioRad, Hercules, CA, USA).
The expression levels of miR-362-3p and JMJD2A mRNA levels were measured by qRT-PCR, which was performed using SYBR Green (Roche, Basel, Switzerland) and ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). All the procedures were performed according to the protocols of manufacturers. The primer sequences were as follows: miR-362-3p forward, 5’-GCCGAGAACACACCTATTCA-3’ and miR-362-3p reverse, 5’-CTCAACTGGTGTCGTGGA-3’; U6 forward, 5’-CTCGCTTCGGCAGCACA-3’ and U6 reverse, 5’-AACGCTTCACGAATTTGCGT-3’; JMJD2A forward, 5’-ATCCCAGTGCTAGGATAATGACC-3’ and JMJD2A reverse, 5’-ACTCTTTTGGAGGAACCCTTG-3’; GAPDH forward, 5’-TGACGCTGGGGCTGGCATTG-3’ and GAPDH reverse, 5’-GCTCTTGCTGGGGCTGGTGG-3’. U6 was used as an internal control gene for miR-362-3p, and GAPDH was used as an internal control gene for JMJD2A mRNA. Relative expression levels were calculated using the 2−ΔΔCt method.15
Cell Proliferation Assay
Cell Counting Kit-8 (CCK-8) assay was used to measure the proliferation of SUNE1 and C666-1 cells. The cells were seeded in 96-well plates (cell density of 2×103 cells/well) and cultured at 37°C in a humidified incubator. When the cells were cultured for 0, 24, 48 and 72 h, CCK-8 reagent was added to each well, and all the cells were further incubated for 2 h. Cell proliferation ability was measured by detecting the optical density (OD) value at 450 nm using a microplate analyzer (Bio-Rad Laboratories, Inc.).
Cell Migration and Invasion Assay
Transwell chambers with a pore size of 8 μm (Corning, Inc.) were used to investigate the migration and invasion of SUNE1 and C666-1 cells. The chambers without Matrigel coating were used for the migration assay, and chambers coated Matrigel were used to conduct the invasion assay. Stably transfected NPC cells (5 × 104 cells/well) were seeded in the upper chamber with serum-free medium. The lower chamber contained medium supplemented with 10% FBS. After incubation at 37°C for 48 h, cells in the lower chamber were stained with 0.1% crystal violet for 20 min at room temperature. Then, under an inverted light microscope (Olympus Corporation, Tokyo, Japan), the cells were counted in five randomly selected fields.
Bioinformatics Analysis and Luciferase Reporter Assay
The starBase v3.0 (http://starbase.sysu.edu.cn/) was used to analyze the expression levels of miR-362-3p in HNSC patients from TCGA and Genomic Data Commons Data Portal databases and predicted the complementary binding sequences of miR-362-3p to JMJD2A. To identify the association of miR-362-3p with JMJD2A, a luciferase reporter assay was performed. The wild-type (WT) and mutant (MUT) of 3’-UTR of JMJD2A were cloned into pmirGLO vector (Promega, USA). JMJD2A-WT and JMJD2A-MUT were then co-transfected into SUNE1 and C666-1 cells with miR-362-3p mimic, miR-362-3p inhibitor, mimic NC or inhibitor NC, respectively, using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. After transfection for 48h, the relative luciferase activity was detected using the dual-luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical Analysis
The statistical analyses were performed using SPSS 22.0 (IBM Corp.) and GraphPad Prism 7.0 software (GraphPad Software, Inc.). Data analyzed in the present study were shown as mean ± standard deviation (SD). In measurement data, the differences between two groups were compared using Student’s t-test, and one-way ANOVA followed by Tukey’s post hoc test was used to compare the differences among multiple groups. Chi-square test was used for comparison between categorical variables. Correlations between different variables were assessed by Pearson correlation analysis. Receiver operating characteristic (ROC) analysis was used to evaluate the ability of miR-362-3p to discriminate NPC patients from healthy controls and to distinguish EBV+ patients from EBV- patients. Kaplan–Meier survival curves and Log rank test were used to analyze the relationship between miR-362-3p expression and overall survival of NPC patients. Cox regression analysis was used to identify the prognostic value of miR-362-3p in NPC patients. All experiments were repeated at least three times. P < 0.05 indicated a statistically significant difference.
Results
Expression of miR-362-3p in NPC Patients
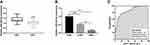
According to the expression data of miR-362-3p in HNSC (497 tumor tissue samples, 44 normal tissue samples) from TCGA and Genomic Data Commons Data Portal databases, miR-362-3p levels were found to be significantly decreased in tumor tissues compared with that in normal tissues (Figure 1A, P < 0.001). In the present study, the significantly downregulated miR-362-3p levels were also found in the serum (Figure 1B, P < 0.001) and tissues (Figure 1C, P < 0.001) of NPC patients compared with healthy serum and normal tissue samples, respectively. Moreover, there was a significant positive correlation between miR-362-3p levels in serum and tissues (r = 0.823, P < 0.001; Figure 1D). An ROC curve was established based on serum miR-362-3p levels in NPC patients and healthy controls to evaluate the diagnostic value of miR-362-3p. The results shown in Figure 1E indicated that serum miR-362-3p had significant diagnostic value for screening NPC patients from healthy controls with an area under the ROC curve (AUC) of 0.915.
Differentially Expressed miR-362-3p Predicts Survival Prognosis in NPC Patients
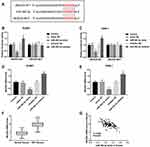
At 5-year follow-up, 41 patients had died, 8 patients were censored, and 63 patients were alive. The expression levels of miR-362-3p in NPC tissues from non-survivors and survivors are shown in Figure 2A, demonstrating that miR-362-3p was significantly decreased in non-survivors compared with that in survivors (P < 0.001). Patients were divided into the high and low miR-362-3p expression group according to the median (1.24) of miR-362-3p expression. Additionally, Kaplan–Meier curves indicated that low miR-362-3p expression was significantly associated with short survival in NPC patients (log-rank P = 0.026, Figure 2B). Moreover, multivariate Cox regression analysis (Table 2) showed that miR-362-3p remained independently associated with patient prognosis under the premise of correcting for patient clinical indicators [hazard ratio (HR) = 2.374, 95% confidence interval (CI) = 1.417–3.338, P = 0.003].
 |
Table 2 Cox Regression Analysis Results for Patients with NPC |
Association of miR-362-3p with the Clinicopathological Characteristics of NPC Patients
As shown in Table 1, no association was observed between miR-362-3p expression and age, gender or smoking (all P > 0.05). However, miR-362-3p expression had a significant correlation with EBV infection (P < 0.001), differentiation (P = 0.013) and TNM stage (P = 0.002). Notably, the significant correlation between miR-362-3p levels and EBV infection suggested that miR-362-3p might be involved in the development of EBV-related NPC.
Different Expression of miR-362-3p in NPC Between EBV+ and EBV- Cases
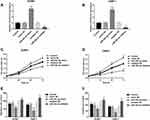
The levels of miR-362-3p were significantly lower in EBV+ patients compared with EBV- patients (Figure 3A, P < 0.001). The results of the analysis of miR-362-3p levels in cells are presented in Figure 3B, miR-362-3p expression was lower in both EBV+ (C666-1) and EBV- (SUNE1) NPC cell lines than that in the normal cell line (all P < 0.001), and the lowest miR-362-3p was observed in EBV+ NPC cell line, and the differences of miR-362-3p expression between EBV+ and EBV- NPC cell lines also reached statistical level (P < 0.01). ROC curve analysis results indicated that miR-362-3p levels had the ability to discriminate between EBV infection positive and negative patients (Figure 3C, AUC = 0.855).
Effects of miR-362-3p on NPC Cell Proliferation, Migration and Invasion
The expression levels of miR-362-3p were markedly upregulated by miR-362-3p mimic and were markedly downregulated by miR-362-3p inhibitor in SUNE1 and C666-1 cells (Figure 4A and B, all P < 0.001). The proliferation of SUNE1 and C666-1 cells was inhibited by miR-362-3p overexpression and promoted by miR-362-3p knockdown (Figure 4C and D, all P < 0.05). The migration (Figure 4E, all P < 0.01) and invasion (Figure 4F, all P < 0.05) of NPC cells (Supplementary Figure S1) were also suppressed by the overexpression of miR-362-3p and promoted by the knockdown of miR-362-3p.
JMJD2A is Directly Inhibited by miR-362-3p in NPC
The complementary binding sequences of miR-362-3p with JMJD2A are presented in Figure 5A. As shown in Figure 5B and C, in SUNE1 and C666-1 cells, the overexpression of miR-362-3p inhibited, and miR-362-3p knockdown facilitated the relative luciferase activity in the JMJD2A-WT group (all P < 0.05), whereas no change was observed in the luciferase activity in JMJD2A-MUT group. In both SUNE1 (Figure 5D) and C666-1 (Figure 5E) cell lines, the mRNA levels of JMJD2A were downregulated by miR-362-3p overexpression and upregulated by miR-362-3p knockdown (all P < 0.01). As shown in Figure 5F, the mRNA levels of JMJD2A were significantly increased in NPC tissues compared with that in normal tissues (P < 0.001). The findings from Figure 5G revealed that JMJD2A mRNA levels in tissues were negatively correlated with the levels of miR-362-3p in tissues (r = −0.564, P < 0.001).
Discussion
Currently, a lot of miRNAs have been reported to be involved in NPC progression, such as miR-101,16 miR-318817 and miR-194.18 In our study, we first found that the expression of miR-362-3p was markedly reduced in HNSC. Then, miR-362-3p expression was found to be reduced in NPC patients, NPC tissues and NPC cells. Aberrant miR-362-3p expression had high diagnostic accuracy in distinguishing NPC patients from healthy individuals. Moreover, the expression levels of miR-362-3p were found to be associated with EBV infection, differentiation and TNM stage. Furthermore, miR-362-3p investigated in the present study is also dysregulated and functions as an important regulator in other cancers. For example, Xu et al have reported that miR-362-3p expression is decreased in glioma tissues and cells and can suppress the development of glioma.19 A study by Wang et al reveals the downregulated expression of miR-362-3p in cervical adenocarcinoma tissues and cell lines, and reveals its inhibitory role in cervical adenocarcinoma.20 In epithelial ovarian cancer (EOC), miR-362-3p levels are low in EOC tissues and cells, and play an anti-proliferative role in EOC.12 Thus, we conclude that miR-362-3p expression may be involved in the progression of NPC.
Considering the potential association between miR-362-3p expression and NPC progression, the prognostic significance of miR-362-3p was then explored. It has been known that miRNAs can function as prognostic biomarkers in various cancers,21,22 including NPC.23,24 In this study, the relative miR-362-3p expression was markedly lower in non-survivors. Besides, patients with low miR-362-3p expression had poor survival compared with the patients with high miR-362-3p expression. The results of Cox regression analysis indicated that miR-362-3p could independently predict survival prognosis in NPC patients. The prognostic role of miR-362-3p has also been shown in other cancers, such as cervical squamous cell carcinoma25 and cervical adenocarcinoma.20 Thus, miR-362-3p can be used as an independent prognostic biomarker for NPC patients.
EBV infection is a high-risk factor for the development of NPC,7 consequently, early screening in EBV+ patients is beneficial to improve the diagnostic accuracy of NPC, provide reasonable treatment promptly, and reduce NPC mortality. The present study showed a significant association of miR-362-3p levels with EBV infection. Then, this study revealed the markedly lower miR-362-3p expression in EBV+ patients than that in EBV- patients, and markedly lower miR-362-3p expression in EBV+ NPC cells than that in EBV- NPC cells. This study also found the ability of miR-362-3p expression to discriminate between EBV+ patients and EBV- patients. In addition, miR-362-3p has been previously reported to be associated with enterovirus 71 and coxsackievirus 16 infection in patients with hand-foot-and-mouth disease (HFMD), and plays a crucial role in distinguishing between infection positive and negative HFMD patients.26 Therefore, it can be speculated that miR-362-3p may be associated with responses such as autoimmune or cellular responses after the organism is infected with the virus, thereby participating in the occurrence and development of EBV infection-related diseases. In clinical practice, the application of miR-362-3p detection may be able to provide a novel diagnostic strategy for the screening of NPC in EBV positive patients.
The biological function of miR-362-3p in NPC progression was then explored. The proliferation, migration and invasion of NPC cells were suppressed by miR-362-3p upregulation and enhanced by miR-362-3p downregulation. Additionally, elevated miR-362-3p has been reported to have an inhibitory effect on cell proliferation, migration and invasion in renal cancer.27 Cao et al have found the suppressive role of miR-362-3p in the function of ovarian cancer cells.11 In breast cancer, miR-362-3p inhibits the cell proliferation.28 Thus, we consider that miR-362-3p serves as a suppressor in NPC progression. Further studies demonstrated that miR-362-3p expression could directly suppress the mRNA levels of JMJD2A in NPC, and mRNA levels of JMJD2A were upregulated in NPC tissues of patients. Thus, miR-362-3p expression may inhibit the proliferation, migration and invasion of NPC cells by targeting JMJD2A. Notably, Su et al have shown that JMJD2A can contribute to the progression of NPC.14 In addition, JMJD2A has also been reported to regulate the cellular function in other cancers, such as bladder cancer29 and cervical cancer.30 Therefore, miR-362-3p may be a suppressor in NPC progression by suppressing JMJD2A.
However, there are still some limitations in the current study. At first, the sample size is relatively small, and further studies with large size are needed. In addition, this study has not elucidated how miR-362-3p levels are regulated following EBV infection, and the mechanism of which urgently needs to be explored by further experimental analysis.
In conclusion, miR-362-3p, which is downregulated in NPC serum, tissues and cell lines, can be used as an independent prognostic biomarker for NPC patients. Additionally, abnormal miR-362-3p is associated with EBV infection in NPC patients, and can be used as a biomarker to screening EBV+ cases from EBV- patients. Moreover, miR-362-3p may be involved in the progression of NPC by suppressing JMJD2A. This study may provide new insights into the pathogenesis of NPC and provide a potential prognostic biomarker and therapeutic target for NPC. In addition, the relationship between miR-326-3p and EBV infection indicates that miR-326-3p may be a biomarker to screen NPC patients with positive EBV infection.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval and Consent to Participate
The experimental procedures were all in accordance with the guideline of the Ethics Committee of Dongying People’s Hospital and have been approved by the Ethics Committee of Dongying People’s Hospital, and this study was conducted in accordance with the Declaration of Helsinki. A signed written informed consent was obtained from each patient.
Consent for Publication
Written informed consent for publication was obtained from each participant.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests.
References
1. Guo R, Mao YP, Tang LL, Chen L, Sun Y, Ma J. The evolution of nasopharyngeal carcinoma staging. Br J Radiol. 2019;92(1102):20190244. doi:10.1259/bjr.20190244
2. Wei KR, Zheng RS, Zhang SW, Liang ZH, Li ZM, Chen WQ. Nasopharyngeal carcinoma incidence and mortality in China, 2013. Chin J Cancer. 2017;36(1):90. doi:10.1186/s40880-017-0257-9
3. Bao L, You B, Shi S, et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene. 2018;37(21):2873–2889. doi:10.1038/s41388-018-0183-6
4. Lin C, Zong J, Lin W, et al. EBV-miR-BART8-3p induces epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma cells through activating NF-kappaB and Erk1/2 pathways. J Exp Clin Cancer Res. 2018;37(1):283. doi:10.1186/s13046-018-0953-6
5. Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomark Prev. 2006;15(10):1765–1777. doi:10.1158/1055-9965.EPI-06-0353
6. Tao Q, Chan AT. Nasopharyngeal carcinoma: molecular pathogenesis and therapeutic developments. Expert Rev Mol Med. 2007;9(12):1–24. doi:10.1017/S1462399407000312
7. Cai L, Long Y, Chong T, et al. EBV-miR-BART7-3p imposes stemness in Nasopharyngeal Carcinoma cells by suppressing SMAD7. Front Genet. 2019;10:939. doi:10.3389/fgene.2019.00939
8. Dong Y, Zhang N, Zhao S, Chen X, Li F, Tao X. miR-221-3p and miR-15b-5p promote cell proliferation and invasion by targeting Axin2 in liver cancer. Oncol Lett. 2019;18(6):6491–6500. doi:10.3892/ol.2019.11056
9. Kang J, Huang X, Dong W, Zhu X, Li M, Cui N. MicroRNA-1269b inhibits gastric cancer development through regulating methyltransferase-like 3 (METTL3). Bioengineered. 2021;12(1):1150–1160. doi:10.1080/21655979.2021.1909951
10. Wang L, Ge S, Zhou F. MicroRNA-487a-3p inhibits the growth and invasiveness of oral squamous cell carcinoma by targeting PPM1A. Bioengineered. 2021;12(1):937–947. doi:10.1080/21655979.2021.1884396
11. Cao S, Li N, Liao X. miR-362-3p acts as a tumor suppressor by targeting SERBP1 in ovarian cancer. J Ovarian Res. 2021;14(1):23. doi:10.1186/s13048-020-00760-2
12. Yuan J, Li T, Yi K, Hou M. The suppressive role of miR-362-3p in epithelial ovarian cancer. Heliyon. 2020;6(7):e04258. doi:10.1016/j.heliyon.2020.e04258
13. Yang S, Zhang X, Sun Y, et al. MicroRNA-362-3p inhibits migration and invasion via targeting BCAP31 in cervical cancer. Front Mol Biosci. 2020;7:107. doi:10.3389/fmolb.2020.00107
14. Su Y, Yu QH, Wang XY, et al. JMJD2A promotes the Warburg effect and nasopharyngeal carcinoma progression by transactivating LDHA expression. BMC Cancer. 2017;17(1):477. doi:10.1186/s12885-017-3473-4
15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
16. Wu RS, Qiu EH, Zhu JJ, Wang JR, Lin HL. MiR-101 promotes nasopharyngeal carcinoma cell apoptosis through inhibiting Ras/Raf/MEK/ERK signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(16):8240. doi:10.26355/eurrev_202008_22580
17. Zhao M, Luo R, Liu Y, et al. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. Nat Commun. 2016;7:11309. doi:10.1038/ncomms11309
18. Yin W, Shi L, Mao Y. MiR-194 regulates nasopharyngeal carcinoma progression by modulating MAP3K3 expression. FEBS Open Bio. 2019;9(1):43–52. doi:10.1002/2211-5463.12545
19. Xu G, Fang P, Chen K, Xu Q, Song Z, Ouyang Z. MicroRNA-362-3p targets PAX3 to inhibit the development of glioma through mediating Wnt/beta-Catenin pathway. Neuroimmunomodulation. 2019;26(3):119–128. doi:10.1159/000499766
20. Wang D, Wang H, Li Y, Li Q. MiR-362-3p functions as a tumor suppressor through targeting MCM5 in cervical adenocarcinoma. Biosci Rep. 2018;38(3):BSR20180668.
21. Wu J, Zhou Z. MicroRNA-432 acts as a prognostic biomarker and an inhibitor of cell proliferation, migration, and invasion in breast cancer. Clin Breast Cancer. 2021;21(4):e462–e470. doi:10.1016/j.clbc.2021.01.014
22. Zhang J, Kong X, Shi Q, Zhao B. MicroRNA-383-5p acts as a potential prognostic biomarker and an inhibitor of tumor cell proliferation, migration, and invasion in breast cancer. Cancer Biomark. 2020;27(4):423–432. doi:10.3233/CBM-190704
23. Ye SB, Zhang H, Cai TT, et al. Exosomal miR-24-3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J Pathol. 2016;240(3):329–340. doi:10.1002/path.4781
24. Hua X, Fan KC. Down-regulation of miR-1181 indicates a dismal prognosis for nasopharyngeal carcinoma and promoted cell proliferation and metastasis by modulating Wnt/beta-catenin signaling. Eur Rev Med Pharmacol Sci. 2019;23(3):1077–1086. doi:10.26355/eurrev_201902_16996
25. Song L, Liu S, Yao H, et al. MiR-362-3p is downregulated by promoter methylation and independently predicts shorter OS of cervical squamous cell carcinoma. Biomed Pharmacother/Biomedecine & Pharmacotherapie. 2019;115:108944. doi:10.1016/j.biopha.2019.108944
26. Cui L, Qi Y, Li H, et al. Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease. PLoS One. 2011;6(11):e27071. doi:10.1371/journal.pone.0027071
27. Zou X, Zhong J, Li J, et al. miR-362-3p targets nemo-like kinase and functions as a tumor suppressor in renal cancer cells. Mol Med Rep. 2016;13(1):994–1002. doi:10.3892/mmr.2015.4632
28. Assiri AA, Mourad N, Shao M, et al. MicroRNA 362-3p reduces hERG-related current and inhibits breast cancer cells proliferation. Cancer Genomics Proteomics. 2019;16(6):433–442. doi:10.21873/cgp.20147
29. Wang F, Li Y, Shan F, et al. Upregulation of JMJD2A promotes migration and invasion in bladder cancer through regulation of SLUG. Oncol Rep. 2019;42(4):1431–1440. doi:10.3892/or.2019.7246
30. Li Y, Wang Y, Xie Z, Hu H. JMJD2A facilitates growth and inhibits apoptosis of cervical cancer cells by downregulating tumor suppressor miR4915p. Mol Med Rep. 2019;19(4):2489–2496. doi:10.3892/mmr.2019.9916
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.