Back to Journals » OncoTargets and Therapy » Volume 10
Aberrant histone modification in CD19+ B cells of the patients with chronic lymphocytic leukemia
Authors Zhou K, Zhang Q, Liu Y, Xiong Y, Wu S, Yang J, Zhou H, Liu X, Wei X, Song Y
Received 2 September 2016
Accepted for publication 9 December 2016
Published 23 February 2017 Volume 2017:10 Pages 1173—1179
DOI https://doi.org/10.2147/OTT.S121301
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Keshu Zhou, Qing Zhang, Yanyan Liu, Yuanyuan Xiong, Shengsheng Wu, Jingke Yang, Hu Zhou, Xinjian Liu, Xudong Wei, Yongping Song
Department of Hematology, Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital, Zhengzhou, People’s Republic of China
Abstract: The aim of this study was to detect the alterations in histone methylation and acetylation in patients with chronic lymphocytic leukemia (CLL). Global histone H3/H4 acetylation and H3K4/H3K9 methylation were detected by the EpiQuik™ global histone H3/H4 acetylation and H3K4/H3K9 methylation assay kits. The mRNA expression of selected chromatin modifier genes was measured by real-time polymerase chain reaction (RT-PCR). Our results found that the global histone H3/H4 hypoacetylation in the CD19+ B cells of patients with CLL (P=0.028 and P=0.03, respectively) and the global histone H3K9 methylation in patients with CLL were significantly increased compared with controls (P=0.02), while there was no significant difference in the global histone H3K4 methylation between the two groups. The level of SIRT1 and EZH2 mRNA expression was upregulated in patients with CLL (P=0.03 and P=0.02, respectively), which increased significantly with progression from Binet stage A to stage C (P=0.015 and P=0.01, respectively) and Rai good to high risk stage (P=0.007 and P=0.008, respectively). The level of HDAC1 and HDAC7 mRNA expression was significantly increased (P=0.02 and P=0.008, respectively) and HDAC2 and P300 mRNA expression was reduced in patients with CLL (P=0.002 and P=0.001, respectively). In conclusion, it is observed that the aberrant histone modification plays an important role in the pathogenesis of CLL.
Keywords: histone methylation, histone acetylation, SIRT1, EZH2, CLL
Introduction
Chronic lymphocytic leukemia (CLL) is a disease characterized by the accumulation of mature, monoclonal B lymphocytes in the peripheral blood, bone marrow and other lymphoid tissues such as spleen, leading to a gradual accumulation of leukemia cells and resulting in cytopenias and/or organomegaly.1 The clinical course of patients with CLL is heterogeneous, ranging from indolent to aggressive, with survival ranging from a couple of months to several decades.2 The mechanisms underlying the aberrant epigenetic changes in CLL are not completely understood, but their effects in the ontogeny of this disease are now well established. As we all known, the growing evidence showed that alterations in chromatin structure are linked to cell cycle regulation, cell death, and overall genome stability. Of the known epigenetic modifications, histone acetylation and histone methylation are the two most documented epigenetic changes.
The histone proteins that package DNA into chromatin play key roles in the regulation of transcription. The N-terminal tails of these proteins are subjected to several posttranslational modifications such as acetylation, deacetylation, and methylation.3 The combination of these covalent modifications gives rise to the so-called “histone code”.4 Transcription becomes active when histones are acetylated by histone acetyltransferases (HATs), silenced when histones are deacetylated by histone deacetylases (HDACs), and silenced or activated when methylated by histone methyltransferases (HMTs).5
The aim of this study was to determine the alterations in global histone H3/H4 methylation/acetylation status and to evaluate the expression profile of HATs, HDACs and HMTs in patients with CLL. Indeed, to our knowledge, no study has correlated expression with the clinical impact of Chinese patients with CLL. Therefore, we carried out the expression analysis of selected members of histone modifier genes: HATs (EP300, CREBBP, PCAF), HDACs (HDAC1, HDAC2, HDAC7, SIRT1), and HMTs (SUV39H1, SUV39H2, EZH2), and further correlated the expression with clinical characteristics.
Patients and methods
Subjects
Peripheral blood samples were collected from 58 newly diagnosed patients with CLL between 2013 and 2015, and 18 healthy controls were enrolled in this study. This study was approved by the ethics committee of the Affiliated Cancer Hospital of Zhengzhou University and conducted according to the principles expressed in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Written informed consent was obtained from all the subjects.
The diagnosis of CLL required a persistent B lymphocytosis of >5.0×109/L and a typical CD19+, CD5+, CD23+, immunoglobulin (Ig) light chain (κ or λ)-restricted immunophenotype as revealed by the flow cytometry of peripheral blood cells, which was according to the guidelines of the International Workshop Chronic Lymphocytic Leukemia/National Cancer Institute (iwCLL/NCI).6 Complete clinical profiles were obtained from all the subjects. All the patients were grouped by age, sex, lactate dehydrogenase (LDH) level, β2 microglobulin (β2-MG) level, Binet stage, and Rai risk stage. High LDH was defined as >245 U/L and β2-MG as >3.5 mg/L.
Cell preparation, RNA isolation, and reverse transcriptase
Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation. CD19+ B cells were isolated by magnetic beads, according to the manufacturer’s instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). The purity of the B cell was >98%, which was detected by the flow cytometry method. Total RNA was isolated by Trizol reagent (Invitrogen), quantified by Nanodrop, and a total of 10 ng of RNA was subsequently used to synthesize complementary DNAs using the Reverse Transcription Kit (Applied Biosystems).
Real-time polymerase chain reaction (RT-PCR)
For PCR amplification, an initial denaturation at 94°C for 10 min was followed by 40 cycles of denaturation at 94°C for 15 s, and extension at 60°C for 1 min. The relative quantification of gene expression was obtained by comparing with the relative expression of β-actin using 2−ΔΔCt method. Each experiment was repeated three times, and the averages were calculated. Selected chromatin modifier genes were used. The primer sequences are shown in Table 1.
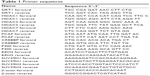  | Table 1 Primer sequences |
Detection of global histone H3/H4 acetylation and global H3K4/H3K9 methylation
Histone extraction and the detection of global histone H3/H4 acetylation and global H3K4/H3K9 methylation were performed using the EpiQuik™ global histone H3/H4 acetylation and H3K4/H3K9 methylation assay kits, according to the manufacturer’s instructions (Epigentek, Brooklyn, NY, USA). In brief, histone proteins (1 mg) were stably spotted on the strip wells. Acetylated histone H3K4/H3K9 was detected with a high-affinity antibody, and the ratios and amounts of acetylated histone H3K4/H3K9 were quantified with a horseradish peroxidase-conjugated secondary antibody color development system. Color was measured by absorbance at 450 nm.
Statistical analysis
SPSS18.0 (SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. The results were presented as mean ± SD. One-way analysis of variance (ANOVA) or the nonparametric Mann–Whitney U-test was used. A P-value <0.05 was considered to be statistically significant.
Results
Patient characteristics
This study included 34 males (60.7%) and 22 females (39.3%) with a median age of 63 years (range, 60–80 years). The median number of white blood cells (WBCs) was 26×109/L (range from 11.5×109/L to 280×109/L). The clinical parameters of 58 patients are summarized in Table 2.
Global histone H3/H4 acetylation and H3K9 methylation in patients with CLL and controls
Histone acetylation and methylation were detected in 15 patients with CLL and 15 healthy controls. Our data showed that the global histone H3/H4 acetylation was significantly decreased in the CD19+ B cells of patients with CLL, when compared with controls (P=0.028 and P=0.03, respectively) as shown in Figure 1A and B. There was no significant difference in the global histone H3K4 methylation between the two groups, as shown in Figure 1C, but the global histone H3K9 methylation in patients with CLL was significantly higher than that of healthy controls (P=0.02), as shown in Figure 1D.
The mRNA expression of chromatin modifier genes in patients with CLL and controls
It was shown that the expression level of SIRT1 and EZH2 mRNA in patients with CLL was significantly higher than that of healthy controls (P=0.03 and P=0.02, respectively), as shown in Figures 2A and 3A. We also investigated whether SIRT1 and EZH2 expression was associated with subgroups of CLL with different clinical characteristics such as Binet stage, Rai risk stage, LDH, β2-MG, ZAP-70, CD38, and FISH cytogenetic. It showed that the level of SIRT1 and EZH2 mRNA expression increased significantly with progression from Binet stage A to stage C (P=0.015 and P=0.01, respectively), as shown in Figures 2B and 3B, and Rai good to high risk stage (P=0.007 and P=0.008, respectively), as shown in Figures 2C and 3C. The level of EZH2 mRNA expression in the β2-MG+ subgroup was increased significantly compared with β2-MG− subgroup (P=0.029), as shown in Figure 3D.
Moreover, it was found that the expression level of HDAC1 and HDAC7 mRNA in patients with CLL were significantly higher than that of healthy controls (P=0.02 and P=0.008, respectively), as shown in Figure 4A and C, while the level of HDAC2 and P300 mRNA expression was reduced significantly in patients with CLL (P=0.002 and P=0.001, respectively), as shown in Figure 4B and D. There was no significant difference in the expression of PCAF, CREBBP, SUV39H1, and SUV39H2 mRNA between patients with CLL and controls, and between any subgroups (Figure 5).
Discussion
CLL is the most common leukemia in adult populations of Western countries. The exact mechanism of the pathogenesis of CLL still has to be determined. In the current study, we focus on the change in histone modification in CD19+ B cells of the patients with CLL.
Our data showed that the global histone H3/H4 acetylation was significantly decreased in the CD19+ B cells of patients with CLL, compared with controls, which was in line with the results of Aron’s data,7 in which depsipeptide, an HDAC inhibition, can increase the histone H3 and H4 acetylation to induce apoptosis in CLL B cells, indicating that the downexpression of histone acetylation involves in the pathogenesis of CLL. SIRT1 is a class III nuclear deacetylase that can activate or repress genetic programs by modifying histones and transcription factors. SIRT1 is upregulated in several human tumor types.8 Interestingly, in this study, it was shown that the expression level of SIRT1 mRNA in patients with CLL was significantly higher than that of controls. Furthermore, the level of SIRT1 mRNA expression increased significantly with progression from Binet stage A to stage C and Rai good to high risk stage; this is keeping with independent observations linking increased expression of the enzyme to tumor transformation.9 Furthermore, it was shown that the expression level of HDAC1 and HDAC7 mRNA in patients with CLL was significantly increased than that of healthy controls, while the level of HDAC2 and P300 mRNA expression was reduced significantly in patients with CLL, suggesting that the abnormal expression of those HDACs may play a vital role in the pathogenesis of CLL.
It was found that the global histone H3K9 methylation in patients with CLL was significantly higher than that of controls, but there was no significant difference in the global histone H3K4 methylation between the two groups, suggesting that the dysregulation of histone methylation plays an important role in the etiology of CLL. EZH2 is overexpressed in several cancer types, both solid tumors and hematopoietic malignancies,10,11 and its overexpression is correlated with disease aggressiveness.12–15 In acute myeloid leukemia, EZH2 inhibited differentiation programs in leukemic stem cells, increasing their leukemogenic activity.14 EZH2 was found to be aberrantly overexpressed in natural killer/T-cell lymphoma (NKTL), conferring a poor prognosis.15 In CLL, it is reported that EZH2 overexpression is related to a poor prognosis of CLL and could be a useful tool to assess its aggressiveness.16,17 In this study, it was found that the expression level of EZH2 mRNA in patients with CLL was significantly higher than that of controls, and EZH2 mRNA expression increased significantly with progression from Binet stage A to stage C and Rai good to high risk stage. The level of EZH2 mRNA expression in the β2-MG+ subgroup was increased significantly compared with β2-MG− subgroup. Those data were in accordance with other’s results,18,19 implicating EZH2 in the pathophysiology of CLL with remarkable clinical and biological heterogeneity. But, the complex mechanisms leading to EZH2 overexpression still have to be better understood.
Conclusion
In this study, the global histone H3/H4 hypoacetylation, H3K9 hypermethylation, and overexpression of SIRT1 and EZH2 were detected in the CD19+ B cells of patients with CLL, indicating that the aberrant histone modifcation was involved in the pathogenesis of CLL.
Acknowledgment
This study was funded by grants from the National Natural Science Foundation of China (81470336).
Disclosure
The authors report no conflicts of interest in this work.
References
Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. | ||
Hamblin T. Chronic lymphocytic leukaemia: one disease or two? Ann Hematol. 2002;81(6):299–303. | ||
Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116(2):259–272. | ||
Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. | ||
Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12(2):142–148. | ||
Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. | ||
Aron JL, Parthun MR, Marcucci G, et al. Depsipeptide (FR901228) induces histone acetylation and inhibition of histone deacetylase in chronic lymphocytic leukemia cells concurrent with activation of caspase 8-mediated apoptosis and down-regulation of c-FLIP protein. Blood. 2003;102(2):652–658. | ||
Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69(5):1702–1705. | ||
Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9(2):123–128. | ||
Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1–2):21–29. | ||
Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106(2):243–247. | ||
Sasaki D, Imaizumi Y, Hasegawa H, et al. Overexpression of enhancer of zeste homolog 2 with trimethylation of lysine 27 on histone H3 in adult T-cell leukemia/lymphoma as a target for epigenetic therapy. Haematologica. 2011;96(5):712–719. | ||
Chen J, Li J, Han Q, et al. Enhancer of zeste homolog 2 is overexpressed and contributes to epigenetic inactivation of p21 and phosphatase and tensin homolog in B-cell acute lymphoblastic leukemia. Exp Biol Med. 2012;237(9):1110–1116. | ||
Tanaka S, Miyagi S, Sashida G, et al. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 2012;120(5):1107–1117. | ||
Yan J, Ng SB, Tay JL, et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood. 2013;121(22):4512–4520. | ||
Papakonstantinou N, Ntoufa S, Chartomatsidou E, Kotta K, Agathangelidis A, Giassafaki L. The histone methyltransferase EZH2 as a novel prosurvival factor in clinically aggressive chronic lymphocytic leukemia. Oncotarget. 2016;7(24):35946–35959. | ||
Rabello Ddo A, Lucena-Araujo AR, Alves-Silva JC, et al. Overexpression of EZH2 associates with a poor prognosis in chronic lymphocytic leukemia. Blood Cells Mol Dis. 2015;54(1):97–102. | ||
Jankowska AM, Makishima H, Tiu RV, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118(14):3932–3941. | ||
Papakonstantinou N, Ntoufa S, Chartomatsidou E, et al. Differential microRNA profiles and their functional implications in different immunogenetic subsets of chronic lymphocytic leukemia. Mol Med. 2013;19:115–123. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






