Back to Journals » OncoTargets and Therapy » Volume 13
A Study of hTERT Promoter Methylation in Circulating Tumour DNAs of Patients with Ovarian Magnificent Tumour
Authors Li S , Huang W, Li Y , Chen B, Li D
Received 30 July 2020
Accepted for publication 24 October 2020
Published 1 December 2020 Volume 2020:13 Pages 12317—12323
DOI https://doi.org/10.2147/OTT.S274743
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Songyi Li, Wei Huang, Yinghua Li, Beibei Chen, Dingheng Li
Department of Gynecology, Hangzhou Women’s Hospital, Hangzhou 310008, People’s Republic of China
Correspondence: Dingheng Li Email [email protected]
Objective: Human telomerase reverse transcriptase (hTERT), a crucial enzyme for telomere maintenance, has been associated with the development of ovarian cancer (OC). The purpose of this study was to investigate the difference of methylation rates of hTERT promoter in tumour tissues and plasma samples of patients with ovarian magnificent tumour and those with ovarian benign tumour, as well as in plasma samples of healthy women. This study further aimed to establish a possible association between increased methylation rate of hTERT promoter and circulating tumour DNAs (ctDNA) amongst patients with ovarian magnificent tumour.
Methods: Tumour tissue samples and plasma samples were separately obtained from 17 patients with ovarian magnificent tumour (experiment group, group A) and from 15 patients with ovarian benign tumour (control group, group B). Another 15 plasma samples were acquired from healthy women (control group, group C). Promoter methylation was assessed by methylation-specific PCR (MSP). Statistical analysis was conducted using SPSS 22.0.
Results: Methylation of hTERT was observed in 76.5% of tumour tissue samples and in 70.6% of plasma samples from patients with ovarian magnificent tumour. It was also observed in 26.7% of tumour tissue samples and 20% of plasma samples from patients with ovarian benign tumour, and in 13.3% of plasma samples from healthy women. Comparing between plasmas and tissues, the respective rates of consistency, sensitivity and specificity were 70.59%, 76.9% and 50% in group A, and 80%, 50% and 90.9% in group B. Hence, the associations of hTERT methylation with ctDNAs (p=0.001) and tumour tissue samples (p=0.012) amongst patients with ovarian magnificent tumour were established.
Conclusion: An increased methylation of hTERT promoter is related to ctDNAs and tumour tissues of patients with ovarian magnificent tumour.
Keywords: human telomerase, promoter, methylation, ovarian tumour
Introduction
The incidence rate of ovarian malignancies ranks third amongst female reproductive system malignancies. Survival of patients with epithelial OC is significantly associated with the stage of disease, thus early detection of OC can substantially increase survival rates.1 However, it is not easy to detect OC in its early stages due to lack of specific detection indicators. Thus, most OC cases have been diagnosed during the middle and late stages. Presently, the typically used tumour markers for OC diagnosis have low sensitivity and specificity. Therefore, determining the specific tumour markers is of great clinical significance for assisting in early OC diagnosis.
About 90% of human cancers can abnormally activate telomerase; relatively, hyper methylation is a usual mechanism for telomerase activation in cancers.2 The hTERT hyper methylation in cancer was once evaluated by analysing normal samples from 1352 tumours and 80 different normal tissues. The median methylation rate of hTERT in normal tissues was 7.0%, with no sample being more than 18.3%. Moreover, 91.4% of tumours had exceeded hTERT methylation levels in normal tissues.3 It was found that the expression of hTERT, as a catalytic protein subunit, was parallel to that of telomerase, which had been highly expressed in OC cases.4 There was methylation of hTERT promoter in several kinds of malignant tumours,5 and its histological fragments could be dissociated from serums.6 It is therefore speculated that hyper methylation of hTERT promoter can be utilised as a prognostic biomarker of ovarian cancer. It has been validated that hTERT can be detected in free DNAs in serums, and the methylation of hTERT promoter can be a predictor of malignant tumours.7,8
Hence, this study aims to verify the significance of hTERT promoter methylation in OC diagnosis.
Materials and Methods
Patients and Samples
In this retrospective study, tumour tissue samples and plasma samples were separately obtained from 17 patients with ovarian magnificent tumour and from 15 patients with ovarian benign cyst. The said patients were surgically treated at Hangzhou Women’s Hospital. Meanwhile, plasma samples were also gathered from 15 healthy women. This study was performed in accordance with the Declaration of Helsinki. All of the enrolled study objects had provided informed consent. The study protocol was approved by the Ethics Committee of the Hangzhou Women’s Hospital. The patients had received no chemotherapy or radiotherapy prior to surgery.
The obtained tumour tissue samples were frozen in liquid nitrogen, and then stored at −80°C for further analysis. DNAs were immediately extracted after the plasma samples were acquired. The median ages at the time of surgical treatment were 44 years old for patients with ovarian magnificent tumour (age range: 32–68) and 42 years old for patients with ovarian benign cyst (age range: 32–68). The median age for the healthy controls was 45 years old (age range: 32–68) (p>0.05). For patients with ovarian magnificent tumour, seven cases were classified as grade I (41.2%), six as grade II (35.3%), and four as grade III (23.5%). There were two histological types: epithelial ovarian cancer (EOC) (15 cases, 88.2%), and immature teratoma (2 cases, 11.8%). Fifteen cases were unilateral (88.2%), and two were bilateral (11.8%). As for patients with ovarian benign tumour, they presented two histological types: epithelial ovarian cyst (11 cases, 73.3%), and mature teratoma (4 cases, 26.7%). Twelve cases were unilateral (80%), and three were bilateral (20%) (Table 1).
 |
Table 1 Distribution of hTERT Promoter Hyper Methylation Relative to Clinical and Pathological Data |
Cell Lines and Cell Culture
According to literature,9 the methylation of hTERT promoter was positive amongst human cervical cancer cells. The cells (HeLa; Shanghai Cell Bank of Chinese Academy of Sciences, China) were cultured at 37°C in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Rockville, MD, USA), and supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin–streptomycin (Gibco) in a humidified incubator that contained 5% CO2.
DNA Extraction and Bisulfite Conversion
Immediately after sectioning, the tissue samples were kept at −80°C. The DNAs were extracted from tumour tissues and HeLa cells by cell/tissue genomic DNA Extraction Kit (No-GK0221, Shanghai Generay Biotech Co., Ltd.). After the blood was immediately drawn, the plasma was isolated by DNeasy Blood and Tissue Kit (No-50214, Qiagen GmbH, Hilden, Germany). Meanwhile, the concentration was measured by NanodropTM 2000 (Thermo Fisher Inc., USA).
MSP
The methylation of the hTERT promoter was determined by MSP. The DNAs were modified by sodium bisulfite using the EpiTect Bisulfite kit (48) Cat (No-59104, Qiagen GmbH, Hilden, Germany). The sensitivity of PCR was determined by FQ PCR. PCR was performed according to the kit instructions of SuperMix I (Beijing TransGen Biotech Co., Ltd.). The reaction mix contained a DNA solution, RNase-free water, bisulfite mix and DNA Protect Buffer, with a total volume of 50μL.
The specific modification steps were as follows: initial denaturation at 95°C for 5 min; incubation at 60°C for 25 min; denaturation at 95°C for 5 min; incubation at 60°C for 85 min; denaturation at 95°C for 5 min; incubation at 60°C for 175 min; and, heat preservation at 20°C. The resulting mixture was eluted to afford the purified DNA. To detect the DNA sequence alterations after bisulfite treatment, MSP was used to distinguish methylated alleles from non-methylated ones. Forward primer 5ʹ- GGAAGTGTTGTAGGGAGGTATTTT −3ʹ and reverse primer 5ʹ- CTTTAACCACTAACCTAATCCAAA −3ʹ (UF) were utilised in the PCR reaction of non-methylated promoter, with a product length of 198 bp. Meanwhile, forward primer 5ʹ- GGAAGTGTTGTAGGGAGGTATTTC −3ʹ and reverse primer 5ʹ- CTTTAACCGCTAACCTAATCCG −3ʹ (MF) were employed in the PCR reaction of methylated promoter, with a product length of 198 bp. The reaction mix of PCR amplification contained 10μL of 2 × TransTaq HiFi PCR Super Mix, 0.5μL of forward primers (10μM), 0.5μL of reverse primers (10μM), 7μL of ddH2O, and 0.2μg of DNA template, with a total volume of 20μL.
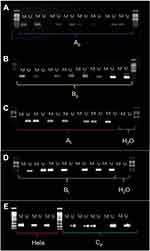
Amplification was performed under the following conditions: initial denaturation at 95°C for 3 min; 40 cycles of denaturation at 95°C for 15 s; annealing at 60°C for 30 s; extension at 72°C for 20 s; and, final extension at 72°C for 7 min. Genomic DNA from the HeLa cells was used as the control of the methylated genes. The PCR products were loaded onto 8% PAA gels, stained with ethidium bromide, and visualised under UV light. The hTERT gene promoter was considered as either: (1) non-methylated, when only the amplification reaction of the non-methylated (U) primers was observed; or, (2) methylated or partially methylated, when the amplification reaction of the methylated (M) primers or both U and M primers was observed. A random reamplification of 20% of the samples was then conducted; no discrepancies were found on the methylation status.
Statistics
Statistical analysis was conducted using SPSS Statistics for Windows Software (Version 22.0). Pearson’s chi-square tests (χ2) were employed for the comparison of different groups. All p values of at most 0.05 were considered statistically significant.
Results
In group A, methylation was detected in tumour tissues (13/17, 76.5%), plasmas (12/17, 70.6%), tissues and plasmas (10/17, 58.8%), and in tissue methylation but not in plasma (3/17, 17.6%, EOC stage I 3cases). Methylation was not detected in tissues, but was detected in plasmas (2/17, 11.8%, EOC stage I 2 case). Comparing plasmas and tissues, the rates of consistency, sensitivity and specificity were 70.59%, 76.9% and 50%, respectively.
In group B, methylation was detected in plasmas (3/15, 20.0%), tumour tissues (4/15, 26.7%), in tissues and plasmas (2/15, 13.3%), in tissues but not in plasmas (2/15, 13.3%), and in plasmas but not in tissues (1/15, 6.7%). Comparing between plasmas and tissues, the rates of consistency, sensitivity and specificity were 80%, 50% and 90.9%, respectively. All of the cases that exhibited positive methylation were epithelial ovarian cysts. The patients’ ages were within 35 to 43 years old, no malignant tumour was found in the general examination, and no abnormality was found after a follow-up period of one year.
In the healthy control group, the hTERT promoter was methylated in two of the 15 plasma samples (13.3%). In the plasma samples, two cases (2/15, 13.3%) were positive for methylation, representing patients with ages 37 and 42 years old. No tumour lesions were found after systemic examination, and no abnormality was found after a follow-up period of one year.
In the HeLa cell group, the hTERT promoter was methylated in all three samples (100%).
The relationship of hTERT methylation amongst the different groups is shown in Table 2. The number of hTERT methylation in tissues and plasmas of the experimental and control groups is depicted in Table 3. The consistency of DNA methylation between the experimental and control groups, as well as the sensitivity and specificity of blood ctDNA detection, are expressed in Table 4. The correlation of hTERT methylation with respect to clinical and pathological features is presented in Table 1. Finally, the MSP images of agarose gel electrophoresis are illustrated in Figure 1.
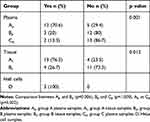 |
Table 2 Methylation of hTERT Relative to the Different Groups |
 |
Table 3 Number of hTERT Methylation in Tissues and Plasmas of the Experimental and Control Groups |
 |
Table 4 Consistency of DNA Methylation Between Tissue Samples and Plasma Samples; Sensitivity and Specificity of Blood ctDNA Detection in the Experimental and Control Groups |
Discussion
Circulating free DNA (cfDNA) is released by necrotic or apoptotic tumour cells. The cfDNA level in cancer patients is higher than that in healthy people. The cfDNA from tumour patients’ plasmas is considered as ctDNA. Some studies have depicted that the positive rate of ctDNA amongst cancer patients is significantly higher than that in non-malignant individuals, and that the quantitative analysis of ctDNA is highly specific (>88%).10–12 In OC, droplet dPCR has a ctDNA detection limit of 0.002%, with high specificity (81%), high sensitivity (99%), and high detection rates (>93%).11 As indicated in some studies, the change of ctDNA is related to the response to adjuvant chemotherapy in OC cases, whilst the level of ctDNA is consistent with the burden of disease and the risk of recurrence amongst OC patients.10,12 Compared to normal epithelial cells, hTERT is overexpressed in over 95% of the OC patients.13 Through detecting methylation of hTERT in plasma ctDNA, tumour cells can be differentiated from normal or benign ones.14 These conclusions have provided relevant evidence for this study, which established the associations of hTERT methylation with ctDNA and tumour tissue samples of patients with ovarian magnificent tumour. However, there have been several cases with benign cysts, and some healthy women have likewise presented methylation of hTERT in plasma ctDNA. These require further research.
Cell-free serum DNA is a suitable and reliable approach for improving early OC detection.15,16 Hyper methylation of the promoter region of ctDNA has been reported to have diagnostic potential.17 As such, ctDNA analysis can be used to determine cancer-specific methylation patterns amongst OC patients. As the primary method for detecting methylated ctDNA in OC patients’ serums,15,18 MSP has been claimed to have a detection limit as low as 0.01%,19 a detection sensitivity of greater than 85%, and a specificity level of greater than 90% for ctDNA considering varied methylation levels.15,17,18 In this study, the hTERT promoter methylation in the ctDNA of OC patients was detected through the MSP method, and was compared with that of patients with ovarian benign tumour as verified on a histological level. The results suggested that the methylation rate of hTERT promoter in the ctDNA of OC patients was significantly higher than that of benign tumour cases and of healthy controls.
The selection of methylation sites in the hTERT promoter region has an influence on the detection results. An analysis of the CPG loci of 92 patients with head and neck cancer (HNSCC) and 53 healthy controls indicated that some sites in HNSCC patients presented a higher frequency of methylation than those in the controls, whilst a contrary finding was observed at another location of CPG.7 By probing the relationship between gene loci and activity of the hTERT promoter, it was found that the first exon, the first intron and the second exon of 37bp in front of the ATG upstream of the promoter of malignant tumour cells were active, whilst their expression in benign cells was relatively low.20 Therefore, in this study, the upstream region of the hTERT core promoter was taken as the methylation detection target area.
The CpG island is located in the hTERT promoter region and is commonly modified by hyper methylation in human cancer cells.3 The methylation of a specific region of the hTERT promoter is identified as a potential biomarker of tumour progression and survival in paediatric gliomas.8 It has been speculated that the methylation of specific regions, especially in the upstream of the hTERT core promoter, can activate the hTERT gene,8 leading to a progression of OC. There are several explanations regarding how the DNA methylation of a hTERT promoter activates hTERT. For one, DNA methylation prevents CTCF, which can inhibit hTERT transcription from binding to the transcription initiation site, thus allowing telomerase activation.21 RIF1 promotes EOC progression by activating hTERT, and the RIF1/hTERT pathway can be a potential therapeutic target for EOC patients.22 Wilms tumour protein is a suppressor of hTERT expression; its binding site depicts an increase of CpG methylation in cancer cells, thus reducing the inhibitory effect of the protein and promoting hTERT expression.23,24 DNA methylation can affect the chromatin conformation change of gene expression,25 which may lead to a binding with different factors and may drive hTERT expressions in cancer cells.26
Hyper methylation is one of the hTERT’s upregulating mechanisms in cancer. Unmethylation is a repressive element of the hTERT promoter. Hyper methylation counteracts this effect, and may regulate other genes proximate to hTERT, which ultimately affect hTERT expression. Hyper methylation as an hTERT-upregulating mechanism, and the understanding of its biological mechanism, require further investigation.3
In this study, the association between hTERT promoter methylation, tumour type and clinical stage was analysed. It was found that the tumour’s type, distribution and clinical stage had no significant correlation. The methylation rate of the hTERT gene promoter in ctDNA and in tumour tissues was greater than that in the control groups, indicating that the hTERT gene promoter methylation was associated with ovarian malignant tumour. Because hTERT promoter hyper methylation is also associated with other epithelial malignancies, the diagnosis of ovarian cancer needs to combine with other tumor markers.
In this study, methylation of hTERT promoter was detected in plasmas and/or tumour tissues of a few patients with benign epithelial ovarian tumours. In addition, there were two cases of positive methylation in the plasmas of healthy controls. However, after further examination and a follow-up period of one year, no malignant tumour was detected amongst these participants. The reason for this requires further determination by methylation gene sequencing.
This study had some deficiencies. There was only one pair of primers amplified in the experiment, thus it was not known whether primers from a different position would have a higher specificity or would achieve a different experimental result. In addition, this study was only a qualitative test, and not a quantitative one. Hence, an accurate quantitative comparison of the primer amplification products was lacking. Since ctDNA methylation detection was non-invasive, easy to sample, repeatable, highly specific and sensitive, hTERT promoter methylation detection was assumed as a potential minimally invasive diagnostic tool for ovarian malignant tumour.
Conclusion
An increased methylation of hTERT promoter is related to ctDNAs and tumour tissues of patients with ovarian magnificent tumour.
Core Tip
In this study, data analysis from clinical sample detection confirmed that the methylation rate of the hTERT DNA promoter in circulating tumour DNAs of patients with ovarian malignant tumour was higher than that of patients with benign ovarian tumour and that of healthy individuals. Meanwhile, there was no significant difference in hTERT promoter methylation between the benign ovarian tumour group and the healthy control group. The methylation rate of the hTERT DNA promoter in ovarian malignant tumour tissues was higher than that in ovarian benign tumour tissues.
Acknowledgment
This work was financially supported by the Fund Project: Medical and Health Science and Technology Project of Zhejiang Province (2017KY553), and by the health commission of Zhejiang province project (2018248116).
Disclosure
The authors declare no conflict of interest in this study.
References
1. Karimi-Zarchi M, Mortazavizadeh SM, Bashardust N, et al. The clinicopathologic characteristics and 5-year survival rate of epithelial ovarian cancer in Yazd, Iran. Electron Physician. 2015;7(6):1399–1406. doi:10.14661/1399.
2. Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448(7155):767–774. doi:10.1038/nature05985
3. Lee DD, Leão R, Komosa M, et al. DNA hypermethylation within TERT promoter upregulates TERT expression in cancer. J Clin Invest. 2019;129(1):223–229. doi:10.1172/JCI121303
4. Maraei AA, Hatta AZ, Shiran MS, Tan GC. Human telomerase reverse transcriptase expression in ovarian tumors. Indian J Pathol Microbiol. 2012;55(2):187–191. doi:10.4103/0377-4929.97865
5. Bougel S, Lhermitte B, Gallagher G, et al. Methylation of the hTERT promoter: a novel cancer biomarker for leptomeningeal metastasis detection in cerebrospinal fluids. Clin Cancer Res. 2013;19(8):2216–2223. doi:10.1158/1078-0432.CCR-12-1246
6. Cristiano S, Leal A, Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570(7761):385–389. doi:10.1038/s41586-019-1272-6.
7. Sobecka A, Blaszczak W, Barczak W, et al. hTERT promoter methylation status in peripheral blood leukocytes as a molecular marker of head and neck cancer progression. J Appl Genet. 2018;59(4):453–461. doi:10.1007/s13353-018-0458-1.
8. Castelo-Branco P, Choufani S, Mack S, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. 2013;14(6):534–542. doi:10.1016/S1470-2045(13)70110-4.
9. de Wilde J, Kooter JM, Overmeer RM, et al. hTERT promoter activity and CpG methylation in HPV-induced carcinogenesis. BMC Cancer. 2010;10(1):271. doi:10.1186/1471-2407-10-271.
10. Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. doi:10.1126/scitranslmed.3003726.
11. Pereira E, Camacho-Vanegas O, Anand S, et al. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One. 2015;10(12):e0145754. doi:10.1371/journal.pone.0145754.
12. Parkinson CA, Gale D, Piskorz AM, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 2016;13(12):e1002198. doi:10.1371/journal.pmed.1002198
13. Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33(5):787–791. doi:10.1016/S0959-8049(97)00062-2.
14. Sirera R
15. Zhang Q, Guohong H, Yang Q, et al. A multiplex methylation-specific PCR assay for the detection of early-stage ovarian cancer using cell-free serum DNA. Gynecol Oncol. 2013;130(1):132–139. doi:10.1016/j.ygyno.2013.04.048
16. Dong R, Yu J, Pu H, et al. Frequent SLIT2 promoter methylation in the serum of patients with ovarian cancer. J Int Med Res. 2012;40(2):681–686. doi:10.1177/147323001204000231
17. Wang B, Lei Y, Luo X, et al. Detection of OPCML methylation, a possible epigenetic marker, from free serum circulating DNA to improve the diagnosis of early-stage ovarian epithelial cancer. Oncol Lett. 2017;14(1):217–223. doi:10.3892/ol.2017.6111
18. Wang B, Lei Y, Yang G-Z, et al. Application of multiplex nested methylated specific PCR in early diagnosis of epithelial ovarian cancer. Asian Pac J Cancer Prev. 2015;16(7):3003–3007. doi:10.7314/apjcp
19. Bondurant AE, Huang Z, Whitaker RS, et al. Quantitative detection of RASSF1A DNA promoter methylation in tumors and serum of patients with serous epithelial ovarian cancer. Gynecol Oncol. 2011;123(3):581–587. doi:10.1016/j.ygyno.2011.08.029
20. cong Y-S, chen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet. 1999;8(1):137–142. doi:10.1093/hmg/8.1.137
21. Renaud S, Loukinov D, Bosman FT, et al. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005;33(21):6850–6860. doi:10.1093/nar/gki989.
22. Liu Y-B, Mei Y, Long J, et al. RIF1 promotes human epithelial ovarian cancer growth and progression via activating human telomerase reverse transcriptase expression. J Exp Clin Cancer Res. 2018;37(1):182. doi:10.1186/s13046-018-0854-8
23. Guilleret I, Yan P, Grange F, et al. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer. 2002;101(4):335–341. doi:10.1002/ijc.10593
24. Shin K-H, Kang MK, Dicterow E, et al. Hypermethylation of the hTERT promoter inhibits the expression of telomerase activity in normal oral fibroblasts and senescent normal oral keratinocytes. Br J Cancer. 2003;89(8):1473–1478. doi:10.1038/sj.bjc.6601291
25. Bert SA, Robinson MD, Strbenac D, et al. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell. 2013;23(1):9–22. doi:10.1016/j.ccr.2012.11006
26. Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi:10.1126/science.1235122
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.