Back to Journals » International Journal of General Medicine » Volume 15
A Seven-Autophagy-Related Long Non-Coding RNA Signature Can Accurately Predict the Prognosis of Patients with Renal Cell Carcinoma
Authors Du R , Xiao Q , Huang J, Feng W, Zheng X, Yi T
Received 4 July 2022
Accepted for publication 1 November 2022
Published 10 November 2022 Volume 2022:15 Pages 8143—8157
DOI https://doi.org/10.2147/IJGM.S381027
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Ruoyang Du,1 Qing Xiao,2 Jianfeng Huang,1 Wubing Feng,1 Xiangqi Zheng,1 Tong Yi1
1Department of Urology, Chongqing Emergency Medical Center, Chongqing University Central Hospital, Chongqing, People’s Republic of China; 2Beijing Ditan Hospital, Capital Medical University, Beijing, People’s Republic of China
Correspondence: Tong Yi, Chongqing Emergency Medical Center, Chongqing University Central Hospital, No. 1, Jiankang Road, Yuzhong District, Chongqing, 400014, People’s Republic of China, Email [email protected]
Introduction: Introduction. Renal cell carcinoma (RCC) is a common malignant tumor worldwide, and to explore, accurate prediction models are essential to the diagnosis and treatment.
Methods: In the present study, the profile expression of RCC patients for long non-coding RNAs (lncRNAs) were obtained from the database of The Cancer Genome Atlas (TCGA). The Gene Set Enrichment Analysis (GSEA) showed that the gene sets related to autophagy are significantly differentially expressed among the paired normal tissues and RCC. Multivariate and univariate Cox analyses were used to construct the gene signature related to prognosis. Receiver Operating Characteristic (ROC) dependent on the time factor and the Kaplan-Meier curves are used for evaluating identified signatures. For gene signature combination, a nomogram with associated clinical constraints was designed.
Results: Multivariate and Univariate Cox analyses presented seven autophagy-related lncRNAs were significantly correlated with Overall Survival (OS) for people with RCC. Risk scores of lncRNA prognostic signature, related to autophagy helped in distinguishing the patients accurately among the low-risk and high-risk RCC based on age, sex, grade of tumor, T, M, N, and AJCC stages. The RCC condition patients, as per their signature were put into the category of low and high-risk groups, having varying prognostic outcomes. Gene signature is an independent prognosticator for OS, accurately predicting 3– 5 year survival time for RCC patients from either of the two groups. GSEA revealed that high-risk scores of the prognostic seven-long non-coding signature correlate with immune-regulatory and cancer, signaling pathways, whereas a low-risk score correlates with metabolism. The quantitative reverse chain reaction of transcription-polymerase verified differential expression of seven lncRNAs autophagy-related samples.
Conclusion: The results depict that seven autophagy-related lncRNA prognostic signature helps in predicting the prognosis accurately among people with RCC.
Keywords: autophagy, lncRNA, renal cell carcinoma, prognosis, GSEA
Introduction
Renal Cell Carcinoma (RCC) is another cancer, developed commonly in human urinary organs, with an incidence rate of 3% of entire body malignant tumors. The incidence of RCC is rising annually by 2% all over the world.1 Despite significant advances in surgery, molecular immunity, gene therapy, and molecular targeted therapy against tumor angiogenesis, patients with RCC present with poor survival chances. On average, the survival time of people with metastatic RCC (mRCC) is only 13 months, and approximately 50% of patients have less than one year of median survival time. In contrast, the 5-year survival time stands at only 10%.2 Therefore, it is crucial to establish tools to predict the prognosis accurately among patients suffering from RCC. It would also guide the clinicians in diagnosing and treat RCC for better therapeutic outcomes.
Autophagy is defined as a conserved evolutionarily catabolic process, in which misfolded proteins, damaged as well as aged organelles, and mutated proteins can be confiscated in double-membrane vesicles called autophagosomes for digestion and degradation in the lysosome.3 However, dysregulation of autophagy is implicated in several human diseases, such as neurodegenerative diseases,4 cardiomyopathies,5 infectious diseases,6 type II diabetes,7 fatty liver disease,8 and cancer.9 Some scholars reveal that Inhibition of PI3K/AKT/mTOR signaling pathway could induce autophagy-mediated cell death in renal cancer.10 There are also studies that inspire us that improve the efficacy of certain drugs on renal tumors by inhibiting p53, and this effect may play a role in inhibiting the autophagic process induced by p53.11 Interestingly, under normal physiological conditions, autophagy prevents the accumulation of proteins and organelles damaged by carcinogens and inhibits malignant transformation. In contrast, the process of autophagy can also provide nutrition to the cancerous cells for promoting higher tumor growth.12 Therefore, it was also essential to discover the biomarkers related to autophagy and develop prognostic signatures, which also serve as the predictive biomarkers and early diagnostic factors for people suffering from RCC.
The Human Genome Project revealed that protein-coding genes account for less than 3% of the total number of human genes, and approximately 90% of the transcribed genes do not encode proteins.13,14 Such transcripts without protein expression are called non-coding RNAs (ncRNAs). In recent years, ncRNAs have gradually become a research hotspot. Long non-coding RNAs (lncRNAs) were other common ncRNAs that can extend up to 200 nucleotides in length. LncRNAs also help in regulating essential functions in biology like the survival, growth of cells, chromatin modifications, genomic imprinting, as well as regulation of allosteric enzyme activities.15 Furthermore, lncRNAs hold a vital part in the pathogenesis of several human diseases, including the development and progression of several types of cancers.16–18 A previous study has shown that lncRNAs regulate autophagic functions; the downregulation of lncRNA MEG3 promotes the proliferation of bladder cancer cells by activating autophagy.19 Some latest discoveries in transcriptome sequencing have enabled the discovery of several lncRNAs related to RCC, as well as novel functions and mechanisms of some specific lncRNAs.20,21 Studies also present the fact that few of the lncRNAs could also act as potential prognostic biomarkers in Hepatocellular Carcinoma (HCC).22,23 Some other researchers also report the potential of lncRNA signatures in the prediction of HCC prognosis.24,25 However, studies on the value of potential lncRNA signatures related to autophagy in predicting survival outcomes for people with RCC are lacking. Therefore, in the present research, using the database of The Cancer Genome Atlas (TCGA), a seven-lncRNA signature is developed with autophagy-related gene sets for predicting the survival time for people having RCC. Moreover, with the help of different cohorts of RCC patients, an inclusive analysis was carried out to validate and identify the robust and novel molecular signature. Furthermore, we investigated prognostic values of molecular signature in different clinical groups and also identified the primary biological process and molecular method involved in tumorigenesis as well as in RCC development using Gene Set Enrichment Analysis (GSEA). Overall, our study aimed to provide valuable information for facilitating more effective stratification and predicting the survival outcomes of people having RCC using lncRNA signatures.
Materials and Methods
Collection of Clinical Samples and Transcriptional Data
Clinical data and RNA expression details for all the considered patients were taken through the database of TCGA-GDC (https://portal.gdc.cancer.gov/). The medical data (Supplementary File S1) extracted from the website includes information about sex, age, Overall Survival (OS), grade, stage, distant metastasis, and metastasis of lymph node. The autophagy-related genes (Supplementary File S2) expression profiles were taken from a database of GSEA (http://www.gsea-msigdb.org/gsea/index.jsp). People with vague living status or incomplete medical data were not included in the study. Tumor tissues were extracted from postoperative specimens of patients in our research institution. This study is consistent with the Declaration of Helsinki. And it was approved by the ethics committee of Chongqing Emergency Medical Center[number 2021–37]. The patients have provided written informed consent and agreed to publish case details.
Autophagy-Related lncRNAs Identification and Prognostic Model Construction
To discover autophagy-related lncRNAs from clinical data, Pearson correlation analysis was carried out. The connection between lncRNAs and autophagy-related genes was made on expression levels. The criteria for Selection were |R|>0.4 and P<0.001 were used to select lncRNAs (Supplementary File S3). Multivariate and Univariate Cox regression analyses were done with the help of the survival package of R, hazard ratios (HRs), β (Cox), and P-values were obtained. HRs are used for identifying and protective lncRNAs (HR<1) and risk-related lncRNAs (HR>1). Afterward, seven target autophagy-related lncRNAs were selected to build a prognostic signature model. The predictive model is made using seven autophagy-related lncRNAs using the following formula:
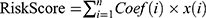 .26 Where Coef (i) is the regression coefficient estimated and x (i) is the expression value of every autophagy-related lncRNA.
.26 Where Coef (i) is the regression coefficient estimated and x (i) is the expression value of every autophagy-related lncRNA.
Verification and Evaluation of the Accuracy of Prognostic Signature
The survival curve Kaplan-Meier was used to assess the survival time for groups with high and low risk. The expression was then drawn out using a heat map and scatter point profiles of autophagy-related genes and the prognosis of patients with RCC in the two groups. Finally, The prediction accuracy was assessed by using the ROC curve. To validate the connection of the risk score with the clinical features of RCC patients, Pearson’s correspondence analysis has been done. The ability of autophagy-related lncRNAs to independently predict the risk of RCC was assessed using multivariate and univariate Cox regression analyses. Additionally, a stratified analysis was carried out to examine the prognostic precision prediction in the survival rates of patients based on other clinicopathological features.
Validation and Establishment of the Nomogram
A nomogram was designed to offer a reliable method to estimate the survival periods of RCC patients for three and five years based on risk values and other clinicopathological characteristics. Next, calibration curves have been used to evaluate the accuracy between the patient survival time anticipated and the observed value. The area underneath the ROC curve (AUC) values for the prediction of RCC prognoses to measure the accuracy of our nomogram.
Gene Set Enrichment Analysis (GSEA)
GSEA (Gene Set Enrichment Analysis) was conducted to discover functional phenotypes between the high-and low-risk groups. The GSEA software was (https://www.gsea-msigdb.org/gsea/index.jsp) used to discover high-risk and low-risk differentially expressed genes. After 1000 permutations, an enriched gene set was achieved on the P-value less than 0.05 with a false discovery rate (FDR) less than 0.25.27 A study of the signals in RCC in high-risk and low-risk groups was conducted in the Kyoto Encyclopedia of Genes and Genomes (KEGG).
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from the tumor and adjacent normal tissues using TRIzol reagent (Invitrogen, California, USA). When these tissues were obtained, none of the patients had received any special treatment other than surgery. The concentration of RNA is estimated using a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, USA). Then, RNA is reverse-transcribed to cDNA with the help of a Prime-Script RT reagent Kit with gDNA Eraser (Takara) according to the manufacturer’s instructions. The lncRNA expression stages are estimated with the help of an SYBR Green PCR Master Mix kit (Takara, Beijing, China) on an Applied Biosystems Real-time 7500 PCR system using GAPDH for internal control. The levels of expression in lncRNAs are quantified relative to those of the internal standard applying the 2–ΔΔCt method. Every sample was measured three times, and the average relative expression was calculated. The primer sequences applied here in the study are written in Table 1.
 |
Table 1 The Primer Sequences Applied in the qRT-PCR |
Statistical Analysis
Data is estimated using a programming language: PERL (https://www.perl.org/), version (Strawberry-Perl-5.32.0.1–64bit.msi). R software (https://www.r-project.org/), version R x64 4.1.0 was used to perform statistical analysis plot graphs.
Results
Acquisition of Prognostically Significant Autophagy-Related lncRNAs in Samples from RCC Patients
Initially, 538 people and their medical data about clear-cell RCC used in this research, was downloaded. Moreover, we extracted the expression profiles of 232 autophagy-related genes through Human Autophagy Database (HADb) and acquired expression profiles of 442 lncRNAs related-autophagy with the help of Pearson correlation analysis among the lncRNA samples and autophagy-related genes as per the following selection standard: |R|>0.4 and P<0.001. Univariate Cox regression analysis in combination with the clinical survival data revealed a significant correlation of 20 lncRNAs with survival time (P < 0.05) (Supplementary File S4). By performing multivariate Cox regression analysis, the following seven potential lncRNAs were filtered out to construct the prognostic signature: AL021707.6, HOTAIRM1, AC084876.1, AC010973.2, LINC01507, AC016773.1, and AC026401.3). Among these, the HRs of HOTAIRM1, AC084876.1, AC016773.1, and AC026401.3 were more significant than 1, while those of the remaining lncRNAs were smaller than 1 (Table 2).
 |
Table 2 By Performing Multivariate Cox Regression Analysis, the Following Seven Autophagy-Related lncRNAs |
Verification of the Accuracy of the Seven-Autophagy-Related-lncRNA Prognostic Signature
The patients were categorized among two groups: the low-risk group (n = 265), and high-risk group (n = 265) as per the corresponding median cut-off values. Patients with RCC were ranked as per the risk scores of seven autophagy-related lncRNAs (Figure 1A). The scatter dot plot showed the connection between RCC survival rates and risk values; the time for the survival of patients with a higher risk scoring was reduced (Figure 1B). Kaplan-Meier Survival Curve Studies showed a much longer survival time and better prognosis for patients in the low-risk group than those in the high-risk (Figure 1C). The ROC curve showed that risk ratings might reliably be used to predict patients’ prognoses. RCC in terms of 1-, 3- and 5-year survival rates, and their AUC values greater than 0.7 (Figure 1D). In the heat map, the difference in expression of the lncRNAs in low and high-risk groups is depicted. The expression of four lncRNAs (HOTAIRM1, AC084876.1, AC016773.1, and AC026401.3) was upregulated in high-risk patients, while that of three lncRNAs, AL021707.6, AC010973.2, and LINC01507, was upregulated in low-risk patients (Figure 1E).
Correlation Analysis of Autophagy-Related lncRNAs Prognosis Signature with Clinicopathological Features
To identify correlations between the clinicopathological characteristics and risk scores of patients, we performed Pearson’s correlation analysis, which showed that except sex (P=0.3395, Figure 2B), other parameters like age (P=0.0465, Figure 2A), grade (P<0.0001, Figure 2C), AJCC (American Joint Committee on Cancer), stage (P<0.0001, Figure 2D), T stage (P<0.0001, Figure 2E), N stage (P<0.0001, Figure 2F), and M stage (P<0.0001, Figure 2G) are significantly associated with risk scores. These findings show that the signature of lncRNA with seven-autophagy is strongly related to the development of RCC.
Assessment of the Possibility of Autophagy-Related lncRNAs to Predict the Risk of RCC
Next, we verified whether autophagic-related signature lncRNA might be used by Cox univariate and multi-variated regression analysis as an independent forecaster in patients with RCC. Univariate analysis showed that T (P<0.001), N (P<0.001), M stage (P < 0.001), AJCC stage (P<0.001), grade (P<0.001), and risk score of autophagy-related prognostic lncRNAs (P<0.001) were correlated significantly to the survival time, but not sex (P=0.951) and age (P=0.134) (Figure 3A). The multivariate analysis resulted that M-stage (P<0.001) and the Risk values based on the lncRNA prognostic signature related to autophagy (P < 0.001) are significantly correlated to the survival time (Figure 3B). In the interim, multiparameter ROC analyses depicted that the AUC value of the risk scores was 0.751 (Figure 3C). Therefore, all the listed findings showed that risk scores based on the lncRNA related-autophagy prognostic signature could predict the prognosis of patients with RCC independently.
Stratification Analysis with Other Clinicopathological Features
A stratification analysis was performed on an entire cohort of people having RCC according to their clinicopathological characteristics, such as tumor grade, age, sex, and AJCC stage. As shown in Figure 4, the survival curve analysis of Kaplan-Meier demonstrated the strong correlation between risk ratings based on autophagic lncRNA signature with OS rates of patients depending on the following features: age (>65 years vs ≤65 years), sex (female vs male), tumor grades (high vs low grade), metastasis (distant metastasis vs no distant metastasis), AJCC stages (early-stage [I–II] or advanced-stage [III–IV] RCC), primary tumor (early primary tumor [stages 1–2] vs advanced primary tumor [stages 3–4]), and lymph node metastasis (regional vs no regional lymph node metastasis). All these findings have revealed the precise prediction of prognosis in various patient groups based on the autophagy-associated lncRNA profile.
Clinical Parameters Quantification and Prognostic Accuracy Evaluation of Risk Score Using a Nomogram
To compute the score for the prediction model accuracy evaluation, a nomogram based on the clinicopathological characteristics and the prognostic autophagic lncRNA signature was used. A nomogram is constructed (Figure 5A) based on multiple clinicopathological traits, like sex, age, grade, AJCC stage, T stage, as well as the risk scores according to seven autophagy-related lncRNAs for accurately estimating 3- and 5-year survival period for the patients with RCC. The calibration study explained the correspondence between the three and five-year OS predicted observed for RCC patients. (Figure 5B and C). Furthermore, the time-dependent ROC revealed that the AUC values were 0.787 for both 3 and 5 years. (Figure 5D). The above outcomes are verified with reliability and accuracy of the risk score based on the seven-autophagy-related-lncRNA signature.
Gene Set Enrichment Analysis (GSEA)
We conducted GSEA comparing high-risk and low-risk groups to determine the possible pathways and their functions associated with the lncRNA signature that participates in RCC development. In the high-risk group, autophagy-related lncRNAs were enriched significantly in immune-related signaling pathways and cancer-related signaling pathways, including autoimmune thyroid disease, primary immunodeficiency, systemic lupus erythematosus, basal cell carcinoma, as well as the P53 signaling pathways (Figure 6A). Additionally the autophagy-related lncRNAs, in the low-risk group, are significantly higher in energy metabolism-related signaling pathways and pathways related to amino acid metabolism, including butanoate, fatty acid, propanoate, sphingolipid, and tryptophan metabolic pathways, which suggests that autophagy-related lncRNA signature participated in the metabolic pathways regulation. GSEA also revealed that the lncRNA autophagy-related signature also participated in the regulation of the classical mTOR signaling pathway (Figure 6B).
Expression Evaluation of Seven Autophagy-Related lncRNAs in Normal Renal and RCC Tissues
To verify the variations in the expression of the seven autophagy-related lncRNAs in normal renal tissues and RCC tissues, we collected both the normal and tumor tissues adjacent to the tumor from 20 patients with RCC and subjected them to qRT-PCR analysis. The results showed that unfavorable factors (HR>1), such as HOTAIRM1, AC084876.1, AC016773.1, and AC026401.3, were increased significantly in RCC (Figure 7A–D). In contrast, the expression of favorable factors (HR<1), AC010973.2 and LINC01507, in RCC tissues was decreased (Figure 7E and F). Among them, the expression of AL021707.6 was not significantly different between normal and RCC tissues (Figure 7G). This result may be attributed to the small sample size of our study, and further follow-up studies were also considered for validating the utility of the prognostic signature.
Discussion
RCC is amongst the highly invasive urinary system tumors. In 2018, 175,098 deaths caused by RCC were reported, which makes it the top 14th cause of fatalities with cancer globally.28 RCC represents one of the major immunogenic carcinomas.29 With its lower sensitivity to chemotherapy and radiotherapy, RCC was systemically treated by radical resection.30 But approximately one-third of patients with RCC display relapse after surgical excision.31,32 Additionally, high-risk patients with RCC have a higher recurrence rate after excision.33 Therefore, it is imperative to develop tools for the detection of high-risk patients at an early stage. This will facilitate the development of new treatment strategies and the stratification of patients according to the risk of recurrence, thereby aiding clinicians in the proper management of patients with RCC.34 However, the biological behavior of RCC is highly variable, and the traditional TNM staging and clinical imaging data are not adequate for evaluating the RCC patient’s prognosis accurately.35,36 Thus, the new biomarkers must be identified urgently for RCC molecular diagnosis that will help guide clinical strategies to improve OS and Disease-Free Survival (DFS) of people having RCC.
Another common belief is that lncRNAs do not encode the proteins directly, however, they regulate gene expression through various signaling pathways to promote or inhibit the progression of multiple cancer types.37,38 Previous studies state the involvement of lncRNAs in different biological processes of cancer, like the damaged DNA, epigenetic regulation, glucose metabolic disorders, immune escape, cell stemness, autophagy, and EMT (Epithelial-Mesenchymal Transition).19,39,40 We believe that lncRNAs can be used as novel biomarkers for predicting the cancer risk along with the survival outcomes of people suffering from cancer.
The prognostic prediction accuracy was analyzed systematically, in the present study for autophagy-related lncRNAs in RCC with the help of statistical and bioinformatics tools. First, we used the TCGA database to identify 20 autophagy-related lncRNAs which are correlated significantly with OC, as evidenced by a univariate Cox regression analysis of autophagy-related expression of lncRNAs among patients with RCC. Then, based on the performance of autophagy-related lncRNAs in the multivariate Cox regression analysis, prognostic characteristics were determined, and finally, seven autophagy-related lncRNAs (AL021707.6, HOTAIRM1, AC084876.1, AC010973.2, LINC01507, AC016773.1, and AC026401.3) were selected.
Autophagy promotes not only cell survival but also cell death. In response to adverse stimuli, cells can degrade pathogenic proteins and damaged organelles via autophagy to recycle energy and escape from apoptosis or necrosis.41 Kimura T’s team has found that autophagy can protect kidneys through suppressing inflammation.42 In contrast, the excessive activation of autophagy in cells leads to the clearance of essential components, causing cell death.43 Previous studies showed that for many kinds of cancers, including RCC in different stages, autophagy plays an essential function.44,45 Other studies have shown that starvation-induced autophagy can promote invasion and metastasis of bladder cancer, and autophagy regulation can affect the sensitivity of tumor cells to chemotherapeutic agents.46 Therefore, exploring autophagy-related biomarkers is of great significance in predicting the progression of the disease and even prognosis in RCC patients. The function of lncRNAs related to autophagy in RCC, however, has not yet been completely explained. The objective of the research was to find the lncRNAs related to autophagy and to investigate their clinical significance in RCC.
The risk score of RCC patients was computed based on expression in the predictive signature for the seven autophagy-related lncRNAs and split into high-risk and low-risk groups based on their average risk score. It was discovered that patients with RCC in the high-risk group had shorter OS than those in the low-risk group. At the same time, the precise autophagy-related lncRNA prognostic signature was confirmed by the ROC curve analysis for RCC patients.
Researchers often use a nomogram, to discover an efficient and accurate clinical instrument for the prediction of the survival of cancer patients.47 Therefore, to verify that the autophagy-related-lncRNA predictions are more reliable to the prognostic prognosis than other conventional clinical factors, we designed a robust nomogram that is based on T, N, M stage, AJCC stage, sex, age, and risk scores (estimated by an autophagy-related lncRNA prognostic signature) improving the prognosis for RCC patients. Calibration plots showed that the nomogram’s three and five-year survival rate was comparable with the actual rate of survival. The seven autophagy-related signatures predicted the OS of patients with RCC correctly, exhibiting a significant potential for clinical applications, including custom prediction and treatment.
A study of GSEA enhancements showed substantial variations between risky and low-risk groups considering the autophagic signal pathways. First, we discovered that metabolism-related signaling pathways and the mTOR pathway were significantly higher in a low-risk group. Being the only core factor for different distinct complexes, including the 1-mTORC1, mTOR complex (mTOR, raptor, and mLST8), complex 2-mTORC2 (mTOR, Rictor, SIN1, and mLST8), and a putative mTOR complex 3 (mTORC3), mTOR plays a vital part in diverse biological process, like in immunity, metabolism, survival, and cell proliferation, promoting anabolic metabolism (eg, amino acid, glucose, lipid, and nucleotide metabolism) and negatively regulating catabolic processes such as autophagy.48–52 mTOR activity is frequently dysregulated in different human cancers, like liver, breast, prostate, lung, and renal carcinomas.53 The immune-related signaling pathways and some cancer-related pathways (basal cell carcinoma and p53 signaling pathways) were significantly high in the high-risk group. As the most mutated tumor suppressor for human malignancies, p53 acts as a tumor suppressor, controlling the expression of more than 2500 downstream cell cycle and cell death target genes.54,55 p53 It impacts a multitude of very different cell functions and constitutes one of the most essential and well-researched tumor suppressors, including the maintenance of genome integrity and fidelity as well as the metabolism and lifespan. Autophagy substantially inhibits p53 signaling pathways in cancer, which otherwise restricts the tumor’s illness to benign disease.56,57 Moreover, p53 activates autophagy. This may be a p53 function control feedback mechanism as an adaptation to metabolic stress.58,59 The field of immunotherapy for RCC is rapidly expanding and remains extraordinarily promising. Over the past decades, significant progress has been made in the development of new immunotherapeutic agents in the form of checkpoint inhibition and vaccines, and these agents are gradually being incorporated into the treatment of mRCC. Some research shows that despite the relatively high rates of response to combinations of PD-1 and/or CTLA-4 axis inhibition in RCC that form the new standard of care, most patients with RCC do not receive durable clinical benefits from these therapies.60–62 So it is urgent to study the relevant in-depth mechanisms of RCC immunotherapy. As some researchers report that the function of immune cells and extracellular matrix molecules are involved in the establishment of the tumor microenvironment of RCC.63 Optimizing treatment dose and schedule, Investigating biomarkers that may affect disease outcomes, and evaluating the combination of different treatment modalities are also critical in maximizing the potential of immunotherapy.64 Further research in this field will allow us to identify better strategies in utilizing immunotherapeutic agents in RCC.
Finally, we verified the difference in the expression of the seven autophagy-related lncRNAs between normal renal and RCC tissues. Among them, the expression of lncRNAs HOTAIRM1, AC084876.1, AC016773.1, and AC026401.3, which were considered unfavorable factors (HR>1), is increased significantly in RCC. In comparison, the lncRNAs AC010973.2 and LINC01507 expression, which were considered favorable factors (HR<1), were significantly decreased in RCC tissues. These findings were consistent with the expected outcomes. However, the expression of lncRNA AL021707.6 was not quite different between normal and RCC tissues. We found this result unexpected and attributed it to the insufficient sample size of the experiment. However, the effect of the high heterogeneity of tumor cells cannot be completely ruled out. Some previous reports have hypothesized that different internal mechanisms or opposite biological processes may function at different stages of cancer.65,66 Therefore, we will use a larger sample size and perform a more in-depth exploration of the specific molecular mechanisms underlying RCC pathogenesis in future studies.
In previous studies, it was revealed that HOTAIRM1 could promote the migration and invasion of glioblastoma cells,67 HOTAIRM1 also can inhibit the proliferation and invasion of lung adenocarcinoma cells through the miR-498/WWOX axis.68 These studies indicated that HOTAIRM1 was positively correlated with the progression of RCC, consistent with our findings. However, there are few related reports about the other six autophagy-related lncRNAs, which are worthy of further exploration in future mechanism studies. At the same time, some similar studies have found the ability of glycolysis-related genes to predict the prognosis of RCC,69 and also found that some genes play an important role in the immune microenvironment.70 To the best of our knowledge, our study is the first to report the prognostic potential of autophagy-related lncRNAs in RCC, and found six rarely reported lncRNAs with great research potential. This novel autophagy-lncRNA prognostic model we established could help in distinguishing the patients accurately among the low-risk and high-risk RCC based on age, sex, grade of tumor, T, M, N, and AJCC stages, and can also accurately predict 3–5 year survival time for RCC patients from either of the two groups. And through GSEA analysis, we found potential therapeutic targets of RCC. This study provides a theoretical basis for the precise diagnosis and treatment of RCC patients.
This research has several restrictions. The first is that the data and clinical data obtained from the TCGA public data repository may be incomplete or partial in this research. Second, Due to a lack of clinical information on RCC subtypes within a database, we did not examine the expression profile of these RCC-related lncRNAs in various RCC subtypes. Finally, additional research into biochemical experiments like the qRT-PCR for larger sample sizes, autophagosome detection, cell cycle analysis, apoptosis assay, and analyses of clinical data is required to confirm the outcomes of the research further.
All in all, we first identified the predictive signature related to autophagy-lncRNA based on seven lncRNAs that can accurately predict the survival results of patients with RCC. To correctly forecast the survival of patients with RCC, we verified the validity of the prognostic nomogram that was generated by integrating the predictive signature with autophagy-related lncRNA and other clinicological traits. A high prognostic risk score based on the lncRNA signature correlated with the activation of immunoregulatory and cancer-related pathways, whereas a low prognostic risk score associated with the activation of various metabolic pathways. Furthermore, the expression profiles of these lncRNAs in clinical samples were consistent with the TCGA database. These findings provide useful insights into the prediction of RCC prognosis and would help develop potential personalized treatments for patients with RCC belonging to different risk groups.
Data Sharing Statement
All data generated or analyzed during the present study are included in this published article and its Supplementary Material files.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by funds from the Chongqing medical scientific research project [grant number 2021MSXM099].
Disclosure
The authors declare that there are no competing interests associated with the manuscript.
References
1. Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi:10.3322/caac.21208
2. Karakiewicz P, Suardi N, Capitanio U, et al. Conditional survival predictions after nephrectomy for renal carcinoma. J Urol. 2009;182(6):2607–2612. doi:10.1016/j.juro.2009.08.084
3. Behrends C, Sowa ME, Gygi SP, et al. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi:10.1038/nature09204
4. Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkinmediated mitophagy that is disrupted by an ALS linked mutation. Proc Natl Acad Sci USA. 2014;111(42):E4439–48. doi:10.1073/pnas.1405752111
5. McLendon PM, Ferguson BS, Osinska H, et al. Tubulin hyperacetylation is adaptive in cardiac proteotoxicity by promoting autophagy. Proc Natl Acad Sci USA. 2014;111(48):E5178–86. doi:10.1073/pnas.1415589111
6. Chen M, Hong MJ, Sun H, et al. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat Med. 2014;20(5):503–510. doi:10.1038/nm.3521
7. Sarparanta J, Garcia-Macia M, Singh R. Autophagy and mitochondria in obesity and type 2 diabetes. Curr Diabetes Rev. 2017;13(4):352–369. doi:10.2174/1573399812666160217122530
8. Mao Y, Yu F, Wang J, et al. autophagy: a new target for nonalcoholic fatty liver disease therapy. Hepat Med. 2016;8:27–37. doi:10.2147/HMER.S98120
9. Amaravadi RK, Lippincott-Schwartz J, Yin XM, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17(4):654–666. doi:10.1158/1078-0432.CCR-10-2634
10. LoPiccolo J, Blumenthal GM, Bernstein WB, et al. Targeting the PI3K/Akt/mTOR pathway: effective combinations and clinical considerations. Drug Resist Updat. 2008;11(1–2):32–50.
11. Warburton HE, Brady M, Vlatkovic N, et al. P53 regulation and function in renal cell carcinoma. CANCER RES. 2005;65(15):6498–6503.
12. Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17(9):528–542. doi:10.1038/nrc.2017.53
13. Derrien T, Johnson R, Bussotti G, et al. The GENCODEv7 catalog of human long non-coding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi:10.1101/gr.132159.111
14. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi:10.1038/nature11233
15. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi:10.1038/nrg.2015.10
16. Yan X, Hu Z, Feng Y, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28(4):529–540. doi:10.1016/j.ccell.2015.09.006
17. Li CH, Chen Y. Insight into the role of long non-coding RNA in cancer development and progression. Int Rev Cell Mol Biol. 2016;326:33–65. doi:10.1016/bs.ircmb.2016.04.001
18. Sanchez Calle A, Kawamura Y, Yamamoto Y, et al. Emerging roles of long non‐coding RNA in cancer. Cancer Prev Res. 2018;109(7):2093–2100. doi:10.1111/cas.13642
19. Ying L, Huang Y, Chen H, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9(3):407–411. doi:10.1039/c2mb25386k
20. Ren S, Peng Z, Mac JH, et al. RNA—seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer—associated long non-coding RNAs and aberrant alternative splicings. Cell Res. 2012;22(5):806–821. doi:10.1038/cr.2012.30
21. Qiao HP, Gao WS, Huo JX, et al. long non—coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. APJCP. 2013;14(2):1077–1082. doi:10.7314/apjcp.2013.14.2.1077
22. DiStefano JK. Long non-coding RNAs in the initiation, progression, and metastasis of hepatocellular carcinoma. Non Cod RNA Res. 2017;2(3–4):129–136. doi:10.1016/j.ncrna.2017.11.001
23. Wei L, Wang X, Lv L, et al. The emerging role of microRNAs and long non-coding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. 2019;18(1):147. doi:10.1186/s12943-019-1086-z
24. Gu JX, Zhang X, Miao RC, et al. Six‐long non‐coding RNA signature predicts recurrence‐free survival in hepatocellular carcinoma. World J Gastroenterol. 2019;25(2):220–232. doi:10.3748/wjg.v25.i2.220
25. Sun Y, Zhang F, Wang L, et al. A five lncRNA signature for prognosis prediction in hepatocellular carcinoma. Mol Med Rep. 2019;19(6):5237–5250. doi:10.3892/mmr.2019.10203
26. Sun Z, Jing C, Xiao C, et al. An autophagy-related long non-coding RNA prognostic signature accurately predicts survival outcomes in bladder urothelial carcinoma patients. Aging. 2020;12(15):15624–15637. doi:10.18632/aging.103718.10.18632/aging.103718
27. Bandyopadhyay S, Mallik S, Mukhopadhyay A. A survey and comparative study of statistical tests for identifying differential expression from microarray data. JIA Bioinform. 2014;11(1):95–115. doi:10.1109/TCBB.2013.147
28. Bray F, Ferlay J, Soerjomataram L, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
29. Heidegger I, Pircher A, Pichler R. Targeting the tumor microenvironment in renal cell cancer biology and therapy. Front Oncol. 2019;9:490. doi:10.3389/fonc.2019.00490
30. Heo J, Park C, Ghosh S, et al. A network meta-analysis of efficacy and safety of first-line and second-line therapies for the management of metastatic renal cell carcinoma. J Clin Pharm Ther. 2021;46(1):35–49. doi:10.1111/jcpt.13282
31. Sawaki M, Shien T, Iwata H. TNM classification of malignant tumors (breast cancer study group). Jpn J Clin Oncol. 2019;49(3):228–231. doi:10.1093/jjco/hyy182
32. Janzen NK, Kim HL, Figlin RA, et al. Surveillance after radical or partial nephrectomy for localized renal cell carcinoma and management of recurrent disease. Urol Clin N Am. 2003;30(4):843–852. doi:10.1016/s0094-0143(03)00056-9
33. Lam JS, Shvarts O, Leppert JT, et al. Postoperative surveillance protocol for patients with localized and locally advanced renal cell carcinoma based on a validated prognostic nomogram and risk group stratification system. J Urol. 2005;174(2):466–472. doi:10.1097/01.ju.0000165572.38887.da
34. Pierorazio P, Johnson M, Patel H, et al. Management of renal masses and localized renal cancer: systematic review and meta-analysis. J Urol. 2016;196(4):989–999. doi:10.1016/j.juro.2016.04.081
35. Lallas C, Trabulsi E, Kaffenberger S, et al. treatment of exophytic renal cancer smaller than 3 cm: surgery versus active surveillance. J Urol. 2015;193(1):16–18. doi:10.1016/j.juro.2014.10.052
36. Zabell J, Demirjian S, Lane B, et al. Predictors of long-term survival after renal cancer surgery. J Urol. 2018;199(2):384–392. doi:10.1016/j.juro.2017.08.096
37. Li Y, Li W, Liang B, et al. Identification of cancer risk lncRNAs and cancer risk pathways regulated by cancer risk lncRNAs based on genome sequencing data in human cancers. Sci Rep. 2016;6:39294. doi:10.1038/srep39294
38. Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, et al. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. 2018;41(6):585–603. doi:10.1007/s13402-018-0406-4
39. Jiang MC, Ni JJ, Cui WY, et al. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9(7):1354–1366.
40. Wang Y, Lu J, Wu Q, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18(1):174. doi:10.1186/s12943-019-1105-0
41. Das G, Shravage BV, Baehrecke EH. Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb Perspect Biol. 2012;4(6):a008813. doi:10.1101/cshperspect.a008813
42. Kimura T, Isaka Y, Yoshimori T. Autophagy and kidney inflammation. Autophagy. 2017;13(6):997–1003. doi:10.1080/15548627.2017.1309485
43. Macintosh RL, Ryan KM. Autophagy in tumour cell death. Semin Cancer Biol. 2013;23(5):344–351. doi:10.1016/j.semcancer.2013.05.006
44. Weiner LM, Lotze MT. Tumor-cell death, autophagy, and immunity. N Engl J Med. 2012;366(12):1156–1158. doi:10.1056/NEJMcibr1114526
45. Wang Z-L, Deng Q, Chong T, et al. Autophagy suppresses the proliferation of renal carcinoma cell. Eur Rev Med Pharmacol Sci. 2018;22(2):343–350. doi:10.26355/eurrev_201801_14178
46. Tong H, Yin H, Hossain MA, et al. Starvation-induced autophagy promotes the invasion and migration of human bladder cancer cells via TGF-β1/Smad3-mediated epithelial-mesenchymal transition activation. J Cell Biochem. 2019;120(4):5118–5127. doi:10.1002/jcb.27788
47. Mir MC, Marchioni M, Zargar H, et al. Nomogram predicting bladder cancer-specific mortality after neoadjuvant chemotherapy and radical cystectomy for muscle-invasive bladder cancer: results of an international consortium. Eur Urol Focus. 2020;5. doi:10.1016/j.euf.2020.07.002
48. Lynch T, Moloughney J, Jacinto E. The mTOR complexes in cancer cell metabolism. In: In Pi3k-Mtor Cancer and Cancer Therapy. New York, NY, USA: Springer; 2016:29–63.
49. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi:10.1016/j.cell.2012.03.017
50. Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signaling crosstalk. Nat Rev Mol Cell Biol. 2014;15(3):155–162. doi:10.1038/nrm3757
51. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169(2):361–371. doi:10.1016/j.cell.2017.03.035
52. Harwood FC, Klein Geltink RI, O’Hara BP, et al. ETV7 is an essential component of a rapamycin-insensitive mTOR complex in cancer. Sci Adv. 2018;4(9):eaar3938. doi:10.1126/sciadv.aar3938
53. Hua H, Kong Q, Zhang H, et al. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12(1):71. doi:10.1186/s13045-019-0754-1
54. Parrales A, Iwakuma T. Targeting oncogenic mutant p53 for cancer therapy. Front Oncol. 2015;5:288. doi:10.3389/fonc.2015.00288
55. Iwakuma T, Lozano G. Crippling p53 activities via knock-in mutations in mouse models. Oncogene. 2007;26(15):2177–2184. doi:10.1038/sj.onc.1210278
56. Guo JY, Xia B, White E. 2013b. autophagy-mediated tumor promotion. Cell. 2013;155(6):1216–1219. doi:10.1016/j.cell.2013.11.019
57. White E. The role for autophagy in cancer. J Clin Invest. 2015;125(1):42–46. doi:10.1172/JCI73941
58. Tasdemir E, Maiuri MC, Galluzzi L, et al. regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10(6):676–687. doi:10.1038/ncb1730
59. Xiao J, Zhang T, Xu D, et al. Fbxl20-mediated vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes Dev. 2015;29(2):184–196. doi:10.1101/gad.252528.114
60. Motzer RJ, Penkov K, Haanen J, et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med. 2019;380:1103–1115. doi:10.1056/NEJMoa1816047
61. Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med. 2019:NEJMoa1816714. doi:10.1056/NEJMoa1816714
62. Motzer RJ, Tannir NM, McDermott DF, et al. Nivolumab plus Ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med. 2018;378:1277–1290. doi:10.1056/NEJMoa1712126
63. Ghatalia P, Gordetsky J, Kuo F, et al. Prognostic impact of immune gene expression signature and tumor infiltrating immune cells in localized clear cell renal cell carcinoma. J Immunother Cancer. 2019;7:139. doi:10.1186/s40425-019-0621-1
64. Postow MA. Managing immune checkpoint-blocking antibody side effects. Am Soc Clin Oncol Educ Book. 2015;76–83. doi:10.14694/EdBook_AM.2015.35.76
65. Baliu-Piqué M, Pandiella A, Ocana AJC. Breast cancer heterogeneity and response to novel therapeutics. Cancers. 2020;12(11):3271. doi:10.3390/cancers12113271
66. Louault K, Li R, DeClerck Y. Cancer-associated fibroblasts: understanding their heterogeneity. Cancers. 2020;12(11):3108. doi:10.3390/cancers12113108
67. Xie P, Xiang L, Chen R, et al. Upregulation of HOTAIRM1 increases migration and invasion by glioblastoma cells. Aging. 2020;13(2):2348–2364. doi:10.18632/aging.202263
68. Chen T-J, Gao F, Yang T, et al. LncRNA HOTAIRM1 inhibits the proliferation and invasion of lung adenocarcinoma cells via the miR-498/WWOX axis. Cancer Manag Res. 2020;12:4379–4390. doi:10.2147/CMAR.S244573
69. Xing Q, Zeng T, Liu S, et al. A novel 10 glycolysis-related genes signature could predict overall survival for clear cell renal cell carcinoma. BMC Cancer. 2021;21(1):381. doi:10.1186/s12885-021-08111-0
70. Fangshi X, Guan Y, Xue L, et al. The effect of a novel glycolysis-related gene signature on progression, prognosis and immune microenvironment of renal cell carcinoma. BMC Cancer. 2020;20(1):1207. doi:10.1186/s12885-020-07702-7
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







