Back to Journals » Cancer Management and Research » Volume 11
A serum piRNA signature as promising non-invasive diagnostic and prognostic biomarkers for colorectal cancer
Authors Qu A, Wang W, Yang Y, Zhang X , Dong Y, Zheng G, Wu Q, Zou M, Du L, Wang Y, Wang C
Received 4 November 2018
Accepted for publication 6 March 2019
Published 29 April 2019 Volume 2019:11 Pages 3703—3720
DOI https://doi.org/10.2147/CMAR.S193266
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Chien-Feng Li
Ailin Qu,1 Wenfei Wang,2,3 Yongmei Yang,4 Xin Zhang,4 Yuhuan Dong,4 Guixi Zheng,4 Qiuyan Wu,4 Mingjin Zou,4 Lutao Du,1 Yunshan Wang,1 Chuanxin Wang1
1Department of Clinical Laboratory, The Second Hospital of Shandong University, Jinan, Shandong 250033, People’s Republic of China; 2Humanistic Medicine Research Center, Qilu Hospital, Shandong University, Jinan, Shandong 250012, People’s Republic of China; 3Humanistic Medicine Research Center, Shandong University, Jinan, Shandong 250012, People’s Republic of China; 4Department of Clinical Laboratory, Qilu Hospital, Shandong University, Jinan, Shandong 250012, People’s Republic of China
Purpose: Piwi-interacting RNAs (piRNAs) are a novel class of small non-coding RNAs, which are not easily degraded but detectable in human body fluids. Recent studies have shown that aberrant piRNA expression is a signature feature across multiple tumor types. However, the expressions of piRNAs in serum of tumor patients and their potential clinical values remain largely unclear.
Patients and methods: High-throughput sequencing was performed to investigate the serum piRNA profiles, followed by evaluations in serum samples of 220 colorectal cancer (CRC) patients and 220 healthy controls using reverse transcription quantitative real-time PCR (RT-qPCR). Biomarker panels including piRNA-based Panel I and carcinoembryonic antigen (CEA)-based Panel II, were developed by logistic regression model, and their diagnostic potentials were compared. Fagan’s nomogram was plotted to promote clinical application.
Results: We identified five differentially expressed serum piRNAs (piR-001311, piR-004153, piR-017723, piR-017724 and piR-020365), which, when combined in the piRNA-based Panel I, outperformed the CEA-based Panel II (P<0.001) and could detect CRC with an area under the receiver operating characteristic curve of 0.867. In addition, Kaplan–Meier analysis showed that patients with low serum piR-017724 level had worse overall survival (OS) and progression-free survival (PFS). In multivariate Cox regression analysis, serum piR-017724 was an independent prognostic factor for OS and PFS (P<0.05).
Conclusion: Our findings suggest serum piRNA expression signatures have potential for use as biomarkers for CRC detection and to predict prognosis at the time of diagnosis.
Keywords: serum piRNA, colorectal cancer, diagnosis, prognosis, nomogram
Introduction
Currently, colorectal cancer (CRC) is the third most commonly diagnosed cancer and accounts for the second leading cause of cancer death globally.1 CRC survival depends mostly on the stage of disease at diagnosis, typically ranging from a 5-year survival rate of 90% for patients diagnosed at the localized stage to 10% for cancer detected at regional or distant stage.2 Early detection is crucial for effective treatment of CRC and patient survival. Currently available modalities for CRC screening include fecal occult blood test (FOBT), stool DNA test, digital rectal examination, flexible sigmoidoscopy and colonoscopy. However, these conventional tests have several disadvantages, such as low sensitivity and specificity, high cost and discomfort to the patients.3 Therefore, it is urgently necessary to develop sensitive, specific, cost-effective and non-invasive methods to complement and improve current CRC screening strategies.
Piwi-interacting RNAs (piRNAs) are a novel class of non-coding RNAs (ncRNAs), which have been of interest primarily due to their essential role in the maintenance of germ cells.4 Only recently, have researchers discovered their presence in mammalian somatic cells, including human cancers.5 It has been demonstrated that piRNAs are expressed across various tissue types in a cancer-specific manner. Deregulated piRNAs have been detected in kidney, thyroid, uterine, bladder, breast, lung, gastric and colon cancer tissues.6 The aberrant piRNA expression is a signature feature with potential diagnostic or prognostic implications across multiple cancer types.7–9 Besides, recent studies have shown that some piRNAs are involved in carcinogenesis and metastasis of several types of cancers, including CRC, providing compelling evidence for the role of these piRNAs in cancer development.10,11 Notably, piRNAs have been reported to be present in human body fluids, including the serum and plasma of healthy individuals, in a markedly stable form.12–14 And most recently, Petra et al have found that circulating piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer.15 In light of these studies, we hypothesized that detection of circulating piRNAs might provide a non-invasive avenue for diagnosis and prognosis of cancers. Nonetheless, until now, the expressions of circulating piRNAs in CRC patients and their potential clinical utilities remain largely unexplored.
In the present study, we systematically and comprehensively screened piRNA profiles in the serum of CRC patients and healthy controls. The diagnostic value of the markedly dysregulated piRNAs in the serum as an additional indicator for CRC was evaluated and compared with that of currently widely used biomarkers. Moreover, the correlation between the expression level of serum piRNAs identified and the survival rate of CRC patients was examined, to evaluate their potential for prognostic prediction.
Material and methods
Ethical statement
All procedures performed in the study involving human participants were in accordance with the ethical standards of the Clinical Research Ethics Committee of Qilu Hospital, Shandong University and the Declaration of Helsinki. Also, this study was approved by Clinical Research Ethics Committee of Qilu Hospital and the experiments were undertaken with the understanding and written consent of each subject.
Study design
The study design consisted of three phases (Figure 1). In the screening phase, the serum samples were pooled from 10 patients with TNM I-II stage (CRC Group A), 10 patients with TNM III-IV stage (CRC Group B) and 10 healthy controls. The samples were then subjected to sequencing to identify piRNAs that were significantly differentially expressed. In the training phase, we firstly tested the expression of candidate piRNAs in an independent cohort from 120 CRC patients and 120 controls, and then constructed piRNA-based and carcinoembryonic antigen (CEA)-based diagnostic panels. A comparison between these two panels was performed. In the validation phase, we evaluated the diagnostic performance of the piRNA-based panel in another independent cohort of 100 CRC patients and 100 healthy controls. Table S1 lists the clinical and demographic characteristics of CRC patients and healthy controls.
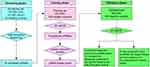 | Figure 1 The flowchart of our study design.Abbreviations: CRC, colorectal cancer; piRNA, Piwi-interacting RNA; RT-qPCR, quantitative real-time PCR. |
Patients and control subjects
A total of 240 blood samples from CRC patients were collected before surgery in the Department of General Surgery of the Qilu Hospital, Shandong University (Jinan, China). None of the CRC patients enrolled had ever received any preoperative therapy (radiotherapy, chemotherapy or chemoradiotherapy) or suffered from other tumor diseases at the same time. Histological specimens from all patients were reviewed to confirm the diagnosis of CRC, and the tumors were staged and graded according to the 2016 TNM classification and the 1973 WHO grading system, respectively. In addition, 230 healthy controls, who visited the hospital for a medical check-up, were enrolled in our study. The healthy controls were screened to ensure that they were within the normal range of all laboratory findings and had no history of cancer. Moreover, the proportions of age and gender in selected healthy controls were similar to those in CRC patients. Furthermore, 20 colorectal adenoma (CRA) patients (12 males, 8 females; age 42–71 years, average 59 years) were included in the study.
The demographic and clinicopathological features of CRC patients, including sex, age, histological differentiation, tumor location, tumor size, tumor stage and preoperative tumor markers [serum CEA and CA19-9], were retrospectively collected. The duration of follow-up was calculated from the date of surgery to death or last follow-up, and the follow-up was completed in December 2016. The overall survival (OS) was defined as the period from time of diagnosis until death. The progression-free survival (PFS) was calculated from the first day of diagnosis to the date when disease progression was detected or the date of death from any cause. The patients were excluded from analyses of clinicopathological characteristics and prognosis if they had incomplete medical records or inadequate follow-up.
Sample collection and preparation
Briefly, 5 mL of whole blood samples were collected from controls and patients by venipuncture. Serum was separated within 1 hr of blood collection by sequential centrifugation (2,000×g for 10 mins at 4°C, 12,000×g for 10 mins at 4°C) to completely remove cell debris. The serum samples were then transferred to RNase/DNase-free tubes and stored at −80°C until further analysis.
Illumina high-throughput sequencing (HTS) and data analysis
The sequencing procedure was performed as previously described.16–18 Briefly, total RNA was isolated from serum using the Trizol LS (Invitrogen, Waltham, MA, USA). The quantity and integrity of RNA yield was assessed using the Qubit2.0 (Life Technologies, Waltham, MA, USA) and Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA) separately. Libraries of small RNAs were constructed with NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (Set 1) (NEB, USA) that is suitable for methylated small RNAs (eg, piRNAs) as well as unmethylated small RNAs, according to manufacturer’s instructions. The libraries were sequenced by HiSeq 2500 (Illumina, USA) with single-end 50nt at Ribobio Co. Ltd (Ribobio, Guangzhou, China).
Raw reads containing adapter or poly ’N’ and reads with low quality and lengths smaller than 17nt were filtered out by FASTQC to get clean reads. Filtered reads were mapped to reference genome by using BWA. Then, mapping reads were matched to pirnabank (
Quantification of piRNAs by reverse transcription quantitative real-time PCR (Rt-qPCR) assays
Briefly, 2 μg of total RNA was reversely transcribed into cDNA using Bulge-Loop piRNA-specific RT primer (Ribobio) and AMV reverse transcriptase (Takara Bio Inc., Shiga, Japan). Then, 2 μL cDNA generated from reverse transcription was used for quantitative PCR analysis that was performed on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) using miScript SYBR Green PCR Kit (Qiagen, Hilden, Germany) and Bulge-Loop primers (Ribobio), based on the previous reported method for piRNA detection.19 The reaction reagents contained 10μL 2XQuantiTect SYBR Green PCR Master Mix, 2μL forward primer of piRNA, 2μL 10XmiScript Universal Primer, 2μL template cDNA. The volume was adjusted with RNA-free H2O. The reaction parameters were incubation at 95°C for 15 mins, then 40 cycles of 95°C for 15 s, 55°C for 30s and 70°C for 30 s. All reactions were performed in triplicate to remove any outliers. In addition, miR-191-5p and U6 were selected as the reference genes according to our previous study.20 The piRNA expression was normalized to the mean of these two reference genes. The relative expressions of piRNAs were determined using the 2−ΔΔCt method.
Statistical analysis
The data distribution of each group was determined by the Kolmogorov–Smirnov test. Continuous variables were presented as the mean ± SD or median (interquartile range) when the values were normally or abnormally distributed, respectively. Statistical differences between the groups were assessed using the Mann–Whitney U-test, unpaired Student’s t-test or a Kruskal–Wallis test as appropriate. Categorical variables were presented as numbers (%) and analyzed using Pearson’s chi-squared test or Fisher’s exact test as appropriate. The Kaplan–Meier method was used to estimate survival, and the survival differences between the subgroups were examined using the log-rank test. A Cox regression model (proportional hazard model) was applied for the univariate analysis and multivariate analysis of prognostic factors. Receiver operating characteristic (ROC) curves were established by MedCalc 9.3.9.0 to discriminate the subjects with or without CRC. Logistic regression analysis was performed using Matlab software (Matlab, R2013a). Fagan’s nomogram was used to help clinicians to calculate a patient’s probability of having CRC according to diagnostic test results. The SPSS (Statistical Package for the Social Sciences) software (version 18.0, Chicago, IL, USA) and R software (version 3.2.3;
Results
Profiling of serum piRNAs in CRC patients by HTS
Analysis of the length distribution showed that all three serum samples contained a number of small RNAs with sizes that were in line with the common size of piRNAs (26–31 nucleotides). Correspondence analysis showed these small RNAs displayed a strong preference for uridine at the 5ʹ end, consistent with the findings of the bias 5ʹ uridine of piRNAs as previously reported by others.21 Among the 23,439 serum piRNAs scanned by Illumina HTS, we chose the piRNAs as potential biomarkers for CRC according to the following criteria: 1) the RPM (the number of reads per million clean tags) values were larger than 1; 2) piRNAs should have at least a twofold difference in expression between the CRC patient group (including CRC Groups A and B) and the control group. As a result, 16 piRNAs were selected based on these criteria for further analysis (Table S2).
Evaluation of piRNAs in serum by RT-qPCR
The expressions of the 16 candidate piRNAs were evaluated by RT-qPCR in serum samples from 120 CRC patients and 120 healthy controls in the training phase. To ensure the stability of the reference genes, we firstly compared the expression levels of miR-191-5p, U6 and their combination in CRC patients and healthy controls. Data showed that there was no significant difference in the expressions of miR-191-5p, U6 and combination of miR-191-5p and U6 between CRC patients and healthy controls (Figure S1A). Among the 16 piRNAs, the expressions of five piRNAs (piR-001311, piR-004153, piR-017723, piR-017724 and piR-020365) were significantly down-regulated in CRC patients compared with the healthy controls (Table 1, Figure S1), with the corresponding area under ROC curve (AUC) values ranging from 0.726 to 0.786 (Figure 2). The detailed sensitivity and specificity of each individual piRNA are summarized in Table S3.
 | Table 1 The selected serum piRNA expression levels in patients with CRC and healthy controls in training set and validation set (median [IQR]) |
We further examined the expressions of these five piRNAs in the validation cohort consisting of 100 CRC patients and 100 healthy controls. There was no significant difference in the distribution of age, gender and tumor characteristic for both CRC and control groups between the training and validation sets. The alterations in the piRNA expression pattern of samples in the validation set were consistent with those in the training set (Table 1 and Figure 3). Moreover, the diagnostic performance of individual piRNAs was explored and Table S3 shows the detailed sensitivity and specificity of piRNAs at the cutoff values determined in the training set (using Youden’s index). The subgroup analysis between colon cancer patients and rectal cancer patients was also performed, but there was no significant difference between both groups (data not shown).
Confirmation of identified piRNAs stability in serum
In order to explore the feasibility of circulating piRNAs as the biomarkers for diagnosis of CRC, we investigated the stability of piR-001311, piR-004153, piR-017723, piR-017724 and piR-020365 in five serum samples by room-temperature incubation and repetitive freeze-thaw cycles. The serum samples were exposed to harsh conditions, including incubation at room temperature for 4, 8 and 24 h, incubation at −80°C for 1, 2 and 3 months, or 1, 2 and 4 repetitive freeze-thaw cycles. The Ct values of these five piRNAs showed no significant variation (Figure S2, all P>0.05), demonstrating that piRNAs were sufficiently stable in serum. Therefore, they could be used for routine processing of clinical samples.
Identification of predictive piRNA panel
A stepwise logistic regression model was constructed to estimate the probability of being diagnosed with CRC in the training set. The predicted risk of being diagnosed with CRC from the logit model based on the five-piRNA-panel (piRNA-based Panel I) was evaluated via the following equation: Logit (P=CRC)=−1.0391+0.1621×piR-001311+0.203×piR-004153+0.3711×piR-017723+0.0897×piR-017724+0.128×piR-020365. We performed ROC curve analysis to assess the diagnostic performance of piRNA-based Panel I, and the results revealed that the AUC for the five-piRNA panel was 0.867 (95% CI: 0.817–0.907) with a sensitivity of 78.3% and a specificity of 74.2% (cutoff value = −0.1085; Figure 2 and Table 2).
 | Table 2 Performance of piRNA-based panel I and CEA-based panel II in the detection of CRC from healthy controls |
Validation of the piRNA panel
The piRNA-based Panel I obtained from the training set was used to predict the probability of being diagnosed with CRC in the validation set. Similar to the training set, the AUC of piRNA-based Panel I was 0.854 (95% CI: 0.797–0.900, Figure S3). We further pooled CRC patients from the training set and validation set, and the combined results showed that the piRNA-based Panel I could detect CRC with an AUC of 0.862 (95% CI: 0.826–0.893) (Table 2). Interestingly, piRNA-based Panel I outperformed the individual piRNAs with regard to CRC detection in all sets (all P<0.05).
In addition, 20 CRA were enrolled in the present study. Figure 3 shows that the expression of piR-017723 was significantly decreased in CRA patients compared with healthy controls. The expressions of the other four piRNAs were not altered in CRA patients compared with healthy controls. Moreover, it seems to be impossible to distinguish CRC from CRA based on the expressions of these piRNAs. The values of piRNA-based Panel I were significantly decreased from the healthy controls through CRA patients to CRC patients (P<0.05).
Comparison of diagnostic potential of the piRNA-based Panel I with CEA and CA19-9
We further investigated the diagnostic roles of CEA and CA19-9 for CRC detection. In the training set, the AUC of the conventional markers were 0.718 and 0.571, and the ROC curves are illustrated in Figure S4. Using logistic regression model, we constructed CEA-based Panel II (including CEA and CA19-9) and plotted its ROC curve (Figure S4C). Then, we compared the AUC values of piRNA-based Panel I and CEA-based Panel II. The results indicate that Panel I showed significantly higher AUC value than Panel II (P=0.0003). When the piRNA-based Panel I was combined with CEA-based Panel II, significant differences were found between the combined model and the piRNA-based Panel I (P=0.0056). Analysis in validation set and combination set produced similar results (Figure 4). The results above confirmed the reliable detective ability of piRNA-based Panel I, which was superior to CEA-based Panel II in detecting CRC to a certain extent.
Construction and assessment of Fagan’ nomogram to calculate the probability of CRC based on piRNA-based panel
The results of combination set showed piRNA-based Panel I could detect CRC with a sensitivity of 0.782 (95% CI: 0.721–0.835) and a specificity of 0.750 (95% CI: 0.687–0.806). To make this information clinically useful, Fagan’s nomogram was established to help clinicians to use Panel I values to estimate the probability of a patient truly having CRC (Figure 5). We set the pre-test probability of CRC at a hypothetical value of 5%. The positive likelihood ratio (+LR) of Panel I was 3.13 and the negative likelihood ratio (-LR) was 0.29. We mark pre-test probability and LR values on the left and middle axes, respectively. Then, a line is drawn from the two marked points along to the right axis. The point at which the line intersects with the right axis is post-test probability. As shown in Figure 5, if a patient had a positive Panel I result, the post-test probability that he truly has CRC would rise to 14.1% (red line). Alternatively, if the patient had a negative result, the post-test probability would decrease to 1.5% (blue line). These data indicate that our five-piRNA based Panel I could provide useful information to clinicians.
 | Figure 5 Fagan’s nomogram for the calculation of the probability that an individual has CRC based on the piRNA-based Panel I.Abbreviations: CRC, colorectal cancer; piRNA, Piwi-interacting RNA. |
Correlation of piRNA expression with survival in CRC patients
We assessed the association of five piRNAs with patients’ survival. A total of 110 patients with complete medical records and adequate follow-up from both training and validation sets were included in the analysis. Kaplan–Meier survival analysis showed that patients with low expression of piR-017724 had significantly lower OS and PFS rate compared with those with high expression of piR-017724 (P=0.005 and P=0.002, respectively, Figure 6). Cox proportional hazards model showed that piR-017724 expression was an independent prognostic factor for OS and PFS in CRC patients (P=0.003 and P=0.004, respectively). The statistical values of piRNAs expression and the other clinicopathological parameters derived from the Cox proportional hazards regression model were listed in Table 3.
 | Table 3 Cox proportional hazards regression model analysis for OS and PFS in CRC patients |
Discussion
Recently, the emergence of circulating ncRNAs as biomarkers has provided a new opportunity to facilitate non-invasive diagnosis and prognosis of cancers. In the present study, we, for the first time, demonstrated a potentially clinically applicable serum piRNA panel that may aid in the development of improved diagnosis and outcome prediction.
The Piwi-piRNA pathway was originally discovered in the Drosophila male germline.22 It is also active in various tumorous cell lines, implying that nonaging somatic cancer stem cells may adopt certain germline-specific characteristics (such as an unlimited proliferation capacity) via the piRNA pathway.23 Piwi-piRNA pathway has been implicated in tumorigenesis via DNA methylation of gene promoter regions, particularly of putative tumor-suppressor genes.24 Additionally, the piRNA expression can delineate clinical features of disease progression.25 For being mostly only 26–31 nucleotide (nt) in length, piRNAs are not easily degraded and can pass through cell membrane easily. This may allow circulating piRNA signatures to be identified in malignant diseases. A new study has revealed that circulating piRNAs are significantly deregulated between colon cancer patients and healthy controls.15 In the present study, we consistently found that piRNAs were differentially expressed in serum from CRC patients compared with controls, and for the first time ever, we constructed a five-piRNA-panel (including piR-001311, piR-004153, piR-017723, piR-017724 and piR-020365) for CRC diagnosis.
Conventionally, CEA and CA19-9 have been used to detect and monitor cancer relapse and prognosis of CRC patients. However, these conventional biomarkers suffer from well-documented disadvantages of limited sensitivity and specificity, especially in case of a single biomarker.26 In our present study, the combined use of these five deregulated piRNAs yielded better diagnostic performance than the use of individual piRNAs. Also, the five-piRNA based Panel I exhibited satisfactory diagnostic performance, showing higher AUC compared with FOBT.27 Furthermore, the diagnostic potential of this five-piRNA based Panel I was better than that of CEA-CA19-9 based Panel II. Since the AUC could only provide global estimation of diagnostic performance, we constructed a Fagan’s nomogram to facilitate the interpretation of test results into clinically useful information.28 With a hypothetical pre-test probability of 5%, the post-test probability of CRC would rise to 14.1% in condition of positive Panel I result, and decrease to 1.5% in condition of negative result.
In addition, we found the scores of the five-piRNA-panel were progressively decreased from the healthy controls through CRA patients to CRC patients. Since adenoma represents a precursor stage of CRC,29 we surmised that decreased panel score in adenomas might be an early indicator in the progression of cancer. Thus, the panel might help identify people with an increased probability of developing CRC in population with familial adenomatous polyposis.
Similar to miRNAs, piRNAs are easier to detect than lncRNA and stable in body fluids. Moreover, piRNAs remain largely undegraded in circulation and are capable of dynamically adapting to a wide range of laboratory conditions.13,14 Considering that stability is crucial for tumor marker, we demonstrated that serum piRNAs remained stable regardless of long-term incubation at room temperature or repetitive freeze–thawing. One possible explanation for such stability can be attributed to that piRNAs are packaged inside exosomes and secreted from cells.30 However, Freedman and colleagues have found that it may not be the case.13 An alternative explanation is that piRNAs may form RNA-protein complexes to maintain stable in the same manner as cell-free miRNAs.31 In this study, the piRNAs were robustly and stably expressed in human blood serum, showing lower Ct values compared with the previously detected miRNAs.32 Therefore, circulating piRNAs possessed the potential as a novel class of blood-based biomarkers for cancer patients.
It is crucial to select suitable reference genes in piRNA research since normalization to inappropriate endogenous control may lead to the identification of incorrect piRNAs of interest. We selected miR-191 and U6, which were used to normalize miRNA expression, as the internal reference genes because they have the advantages of stability and high abundance.20 In this study, we confirmed that there was no significant difference in the mean of miR-191-5p and U6 between CRC patients and healthy controls, indicating that they were also suitable reference genes for piRNA study. Based on this, differentially expressed serum piRNAs were identified in CRC patients by RT-qPCR.
Biomarkers that can identify CRC patients with high risk and poor prognosis are one of the most pressing needs for clinicians. piRNAs have potential functions in transposon silencing as well as epigenetic and post-transcriptional regulation of gene expression.23 Although only a few studies suggest a role for piRNAs in carcinogenesis and progression of cancers,10,11,24,33–35 this newly emerging research topic will surely grow immensely in the near future. Previously, a three-piRNA-panel has been identified in gastric cancer tissue to effectively separate cancer patients by risk of recurrence, which also showed a similar association with recurrence-free survival has been observed in patients with colon cancer.36 In the present study, we investigated the roles of these five identified piRNAs as prognostic biomarkers. Our data demonstrated that the reduced expression of serum piR-017724 was correlated with poor OS and PFS rate. Cox regression model analysis showed that serum piR-017724 was an independent prognostic factor for CRC, suggesting its potential as biomarker for CRC prognosis. Another interesting avenue for future research would be to investigate the piRNA expression in cancer tissues, since research has shown that aberrant expression of piRNAs in tumors has functional consequences.10
Conclusion
This study revealed a novel serum five-piRNA-panel (piR-001311, piR-004153, piR-017723, piR-017724 and piR-020365) for tumor detection and meanwhile, serum piR-017724 was identified as an independent prognosis predictor in patients with CRC.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (Nos 81472025, 81772271, 81601846, 81702084, 81301506, 81874040, and 81501819), Shandong Technological Development Project (2016CYJS01A02 and 2016GSF201120), Taishan Scholar Program of Shandong Province, Natural Science of Basic Scientific Research Foundation of Shandong University (2017BTS01), Science Foundation of Qilu Hospital of Shandong University (2015QLMS51 and 2015QLMS45), Fundamental Research Funds of Shandong University (2014QLKY03), and Key Research and Development Projects of Shandong Province (2015GSF118167).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Du F, Shi SS, Sun YK, Wang JW, Chi Y. Clinicopathological characteristics and prognosis of colorectal cancer in Chinese adolescent patients. Chin Med J (Engl). 2015;128(23):3149–3152. doi:10.4103/0366-6999.170256
3. Barzi A, Lenz HJ, Quinn DI, Sadeghi S. Comparative effectiveness of screening strategies for colorectal cancer. Cancer Am Cancer Soc. 2017;123(9):1516–1527.
4. Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. doi:10.1038/nature04916
5. Cheng J, Guo J, Xiao B, et al. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412(17–18):1621–1625. doi:10.1016/j.cca.2011.05.015
6. Martinez VD, Vucic EA, Thu KL, et al. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep. 2015;5:10423. doi:10.1038/srep10423
7. Ng KW, Anderson C, Marshall EA, et al. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol Cancer. 2016;15:5. doi:10.1186/s12943-016-0491-9
8. Assumpcao CB, Calcagno DQ, Araujo TM, et al. The role of piRNA and its potential clinical implications in cancer. Epigenomics-Uk. 2015;7(6):975–984. doi:10.2217/epi.15.37
9. Han YN, Li Y, Xia SQ, Zhang YY, Zheng JH, Li W. PIWI proteins and PIWI-interacting RNA: emerging roles in cancer. Cell Physiol Biochem. 2017;44(1):1–20. doi:10.1159/000484541
10. Yin J, Jiang XY, Qi W, et al. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017;108(9):1746–1756. doi:10.1111/cas.13300
11. Weng W, Liu N, Toiyama Y, et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer. 2018;17(1):16. doi:10.1186/s12943-018-0767-3
12. Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61(1):221–230. doi:10.1373/clinchem.2014.230433
13. Freedman JE, Gerstein M, Mick E, et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun. 2016;7:11106. doi:10.1038/ncomms11106
14. Yang X, Cheng Y, Lu Q, Wei J, Yang H, Gu M. Detection of stably expressed piRNAs in human blood. Int J Clin Exp Med. 2015;8(8):13353–13358.
15. Vychytilova-Faltejskova P, Stitkovcova K, Radova L, et al. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol Biomarkers Prev. 2018;27:1019–1028. doi:10.1158/1055-9965.EPI-18-0318
16. Vasiliauskaite L, Vitsios D, Berrens RV, et al. A MILI-independent piRNA biogenesis pathway empowers partial germline reprogramming. Nat Struct Mol Biol. 2017;24(7):604–606. doi:10.1038/nsmb.3413
17. Hong Y, Wang C, Fu Z, et al. Systematic characterization of seminal plasma piRNAs as molecular biomarkers for male infertility. Sci Rep. 2016;6:24229. doi:10.1038/srep24229
18. Rubio M, Bustamante M, Hernandez-Ferrer C, et al. Circulating miRNAs, isomiRs and small RNA clusters in human plasma and breast milk. PLoS One. 2018;13(3):e193527. doi:10.1371/journal.pone.0193527
19. Ye W, Liu X, Guo J, et al. piRNA-3878 targets P450 (CpCYP307B1) to regulate pyrethroid resistance in culex pipiens pallens. Parasitol Res. 2017;116(9):2489–2497. doi:10.1007/s00436-017-5554-3
20. Zheng G, Wang H, Zhang X, et al. Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS One. 2013;8(12):e83025. doi:10.1371/journal.pone.0083025
21. Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–258. doi:10.1038/nrm3089
22. Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–764. doi:10.1126/science.1146484
23. Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505(7483):353–359. doi:10.1038/nature12987
24. Yan H, Wu QL, Sun CY, et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29(1):196–206. doi:10.1038/leu.2014.135
25. Huang G, Hu H, Xue X, et al. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15(7):563–568. doi:10.1007/s12094-012-0966-0
26. Tan E, Gouvas N, Nicholls RJ, Ziprin P, Xynos E, Tekkis PP. Diagnostic precision of carcinoembryonic antigen in the detection of recurrence of colorectal cancer. Surg Oncol. 2009;18(1):15–24. doi:10.1016/j.suronc.2008.05.008
27. Niv Y, Sperber AD. Sensitivity, specificity, and predictive value of fecal occult blood testing (Hemoccult II) for colorectal neoplasia in symptomatic patients: a prospective study with total colonoscopy. Am J Gastroenterol. 1995;90(11):1974–1977.
28. Fagan TJ. Letter: nomogram for Bayes theorem. N Engl J Med. 1975;293(5):257. doi:10.1056/NEJM197507312930513
29. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767.
30. Lasser C. Exosomes in diagnostic and therapeutic applications: biomarker, vaccine and RNA interference delivery vehicle. Expert Opin Biol Ther. 2015;15(1):103–117. doi:10.1517/14712598.2015.977250
31. Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi:10.1073/pnas.1019055108
32. Qu A, Yang Y, Zhang X, et al. Development of a preoperative prediction nomogram for lymph node metastasis in colorectal cancer based on a novel serum miRNA signature and CT scans. Ebiomedicine. 2018;37:125–133. doi:10.1016/j.ebiom.2018.09.052
33. Li D, Luo Y, Gao Y, et al. piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int J Mol Med. 2016;38(3):927–936. doi:10.3892/ijmm.2016.2671
34. Li Y, Wu X, Gao H, et al. Piwi-interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol Med. 2015;21:381–388. doi:10.2119/molmed.2014.00203
35. Fu A, Jacobs DI, Hoffman AE, Zheng T, Zhu Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. 2015;36(10):1094–1102. doi:10.1093/carcin/bgv105
36. Martinez VD, Enfield KS, Rowbotham DA, Lam WL. An atlas of gastric PIWI-interacting RNA transcriptomes and their utility for identifying signatures of gastric cancer recurrence. Gastric Cancer. 2016;19(2):660–665. doi:10.1007/s10120-015-0487-y
Supplementary materials
 | Table S1 Characteristics of participants enrolled in the study |
 | Table S2 Selected piRNAs that were significantly downregulated in serum samples of CRC patients compared with healthy controls.The values showing in the columns referred to RPM value |
 | Table S3 Diagnostic performance of individual biomarkers in the training, validation and combination sets |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.