Back to Journals » International Journal of Nanomedicine » Volume 14
A reliable approach for assessing size-dependent effects of silica nanoparticles on cellular internalization behavior and cytotoxic mechanisms
Authors Kim W, Kim WK, Lee K, Son MJ, Kwak M, Chang WS , Min JK, Song NW, Lee J, Bae KH
Received 22 July 2019
Accepted for publication 18 August 2019
Published 10 September 2019 Volume 2019:14 Pages 7375—7387
DOI https://doi.org/10.2147/IJN.S224183
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Wooil Kim, 1,* Won Kon Kim, 1,* Kyungmin Lee, 1 Min Jeong Son, 1 Minjeong Kwak, 2 Won Seok Chang, 3 Jeong-Ki Min, 1 Nam Woong Song, 2 Jangwook Lee, 1 Kwang-Hee Bae 1
1Division of Biomedical Research, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Daejeon 34141, Republic of Korea; 2Center for Nano-Bio Measurement, Korea Research Institute of Standards and Science (KRISS), Daejeon 34113, Republic of Korea; 3Department of Nanoprocess, Korea Institute of Machinery & Materials (KIMM), Daejeon 34103, Republic of Korea
*These authors contributed equally to this work
Correspondence: Jangwook Lee;Kwang-Hee Bae
Division of Biomedical Research, Korea Research Institute of Bioscience and Biotechnology, 125 Gwahak-ro, Yusung-gu, Daejeon 34141, Republic of Korea
Tel +82 42 860 4268
Fax +82 42 860 4149
Email [email protected]; [email protected]
Background: The size of nanoparticles is considered to influence their toxicity, as smaller-sized nanoparticles should more easily penetrate the cell and exert toxic effects. However, conflicting results and unstandardized methodology have resulted in controversy of these size-dependent effects. Here, we introduce a unique approach to study such size-dependent effects of nanoparticles and present evidence that reliably supports this general assumption along with elucidation of the underlying cytotoxic mechanism.
Methods: We prepared and physically characterized size-controlled (20– 50 nm) monodispersed silica nanoparticles (SNPs) in aqueous suspensions. Then, a variety of biochemical assessments are used for evaluating the cytotoxic mechanisms.
Results: SNP treatment in three cell lines decreased cell viability and migration ability, while ROS production increased in dose- and size-dependent manners, with SNPs < 30 nm showing the greatest effects. 30- and 40-nm SNPs were observed similar to these biological activities of 20- and 50-nm, respectively. Under the conventionally used serum-free conditions, both 20-nm and 50-nm SNPs at the IC 50 values (75.2 and 175.2 μg/mL) induced apoptosis and necrosis in HepG2 cells, whereas necrosis was more rapid with the smaller SNPs. Inhibiting endocytosis impeded the internalization of the 50-nm but not the 20-nm SNPs. However, agglomeration following serum exposure increased the size of the 20-nm SNPs to approximately 50 nm, preventing their internalization and cell membrane damage without necrosis. Thus, 20-nm and 50-nm SNPs show different modes of cellular uptake, with smaller SNPs capable of trafficking into the cells in an endocytosis-independent manner. This approach of using non-overlapping size classes of SNPs under the same dose, along with serum-induced agglomeration analysis clarifies this long-standing question about the safety of small SNPs.
Conclusion: Our results highlight the need to revise safety guidelines to account for this demonstrated size-dependent cytotoxicity under serum-free conditions, which may be similar to the microenvironment after tissue penetration.
Keywords: silica nanoparticles, size-dependent cytotoxicity, cellular internalization, necroptosis, serum agglomeration
Corrigendum for this paper has been published
Introduction
Nanoparticles are defined as particles between 1 and 100 nm in size, and their properties significantly differ from those observed with fine particles or bulk materials with the same chemical composition. Whereas bulk materials exhibit consistent physical properties regardless of their size, nanoparticles often show size-dependent properties. Due to their unique characteristics, nanoparticles have great potential for applications in various fields, including biomedical, optical, and electronic applications.1 However, nanoparticles show toxic effects with biological systems above a certain threshold level because of their unusual bioactivities.2 Furthermore, the detailed mechanisms underlying nanoparticle toxicity remain relatively unknown. Thus, a deeper understanding of nanoparticle toxicity would be very valuable for guiding the design of safer nanoparticles and nanomaterials.
Silica nanoparticles (SNPs) have attracted considerable attention and have been used in applications in various fields due to their unique properties, including a large surface area and good biocompatibility. SNPs have been used extensively in applications for chemical mechanical polishing and as additives to drugs, cosmetics, printer toners, and foodstuffs.3,4 Despite these applications, however, the potential risks of SNPs against human heath have not been fully assessed. Recently, SNPs have been widely used for the targeted delivery of contrast agents and drugs and biomedical applications such as biosensors, microscopic imaging, DNA delivery, and enzyme immobilization, in order to improve disease diagnosis and therapy.5,6 The sufficiently small size of SNPs, like that of other nanoparticles, can penetrate relatively large pores of blood vessels around diseased regions, such as in cancer.7 After SNPs are administered to target organisms and cells, they inevitably contact numerous surrounding biomolecules. Therefore, monitoring and understanding the mechanisms associated with cellular uptake, retention, cytotoxicity, and cellular interactions of SNPs deposited in various tissues and organs are of great interest.
Many studies have been conducted in attempt to study the intrinsic properties of SNPs (ie, their sizes, shapes, and surface modifications) and to reveal the mechanisms underlying their toxic effects.8 Particularly in-depth research on the biological responses to SNP size has been performed. Most studies have been performed in the presence of serum, which can aggregate SNPs, showing that smaller SNPs usually exhibit stronger toxicity.9 The smaller the SNP size, the more efficiently they are delivered into cells, suggesting the potential for size-dependent toxicity. However, some conflicting results have been reported regarding the relationship between cytotoxicity and SNP size in studies with SNPs around 50 nm in diameter. Most nanoparticles, including SNPs, are susceptible to aggregation or agglomeration due to serum proteins, making it difficult to maintain the initial synthesized size. Furthermore, in vitro cytotoxicity tests have been performed using SNPs that are not strictly size-controlled, ie, where some overlap occurs between their sizes. Therefore, the cytotoxicity and mechanism of action of SNPs with a well-defined size under agglomeration-free conditions remain unclear.
The induction of oxidative stress, inflammation, and autophagy, leading to apoptotic and/or necrotic cell death, has been reported in various cell lines exposed to SNPs.10 However, there are biases towards toxic mechanisms induced by SNPs internalized into cells. Studies on the endocytic mechanisms and cellular transport of engineered nanoparticles have been well documented.11,12 Previous studies have been performed in attempt to elucidate the size-dependent biological response of SNPs, which focused on toxic effects related to the degree of cellular internalization, rather than on the cellular internalization mechanism itself. It has been reported that SNPs of a certain size or smaller can penetrate cell membranes directly and enter cells.13 Such SNPs would be expected to show different toxicity mechanisms versus SNPs internalized by endocytosis, but the biological responses and toxic effects of membrane-permeable SNPs remain poorly understood. In addition, few studies have been conducted on the effect of serum agglomeration on SNP membrane permeability. Thus, detailed in vitro toxicity studies based on the effects of SNP size and serum on cellular internalization should be taken into consideration.
Here, we prepared size-controlled SNPs in aqueous suspensions that were mono-disperse (20–50 nm) and well dispersed, exhibiting hydrodynamic diameters that compared well with those observed by scanning electron microscopy (SEM). Generally, the size-dependent effects of SNPs on various biological activities, such as cell viability, reactive oxygen species (ROS) generation, genotoxicity, and changes in cellular morphology and motility, were examined in human hepatoma (HepG2), human alveolar carcinoma (A549), and human colon adenocarcinoma (SW480) cells. Although nanoparticles entering the human body can generally be removed by renal or reticuloendothelial system (RES) clearance, significant amounts of nanoparticles are deposited in the liver. Since inorganic nanoparticles like SNPs are not biodegradable, the in vitro toxicity of hepatocytes should be examined more closely than for other cell types. Therefore, prior to studying the detailed cytotoxic mechanisms, we first evaluated the effects on HepG2 cells among the three human cell lines in more detail. Thereby, we treated SNPs with HepG2 cells and reliably examined the size-dependent effects on inducing apoptosis and necrosis in time manner. It is noteworthy that cells treated with 20-nm SNP underwent significant early necrotic cell death. We next prepared FITC-labeled SNPs and closely assessed to their internalization behaviors via FACS and fluorescent microscopy analyses. Finally, it was investigated whether serum proteins affects cellular internalization of 20-nm SNP and the resulting early necrosis.
Materials and methods
Synthesis and characterization of SNPs with different sizes
Spherical SNPs (20-, 30-, 40- and 50-nm in diameter) with narrow size distribution (standard deviation of individual particle size lower than 3.2 nm) were prepared by using modified methods from the previously proposed protocols by Hartlen et al.14 The synthesis of 20-nm SNP was performed in a 500 mL round-bottom flask with 350 mL of 8.2 mM aqueous L-arginine (Sigma-Aldrich) solution. Once the solution was warmed up to 50 °C, the mixture of TEOS (tetraethylorthosilicate, 98%, ACROS) and cyclohexane (99%; Samchun Chemicals) was added while magnetic stirring. The stirring was maintained for 24 h with a fixed speed of 900 rpm to ensure proper mixing between organic phase (TEOS) and water. The reaction resulted in the synthesis of 20-nm spherical SNPs in aqueous suspension with 13 mg/mL in mass concentration.
Procedure for 30-nm SNP synthesis was similar with that for 20-nm SNP synthesis, with a small variation in the mixture concentrations. The mixture of 3.3 mL TEOS and 2.7 mL cyclohexane was added into 41.4 mL of 2.2 mM aqueous L-arginine solution at 70 °C in a 100 mL round-bottom flask and maintained for 24 h. To synthesis spherical 40- and 50-nm SNPs exhibiting suspension stability without agglomeration, regrowth process was adopted using 30-nm SNPs as seeds for further growth. For this regrowth of SNPs up to 40- and 50-nm in size, respectively, an additional volume of TEOS was added into the vessel for 30-nm SNP synthesis after each 24 h. These reactions were performed sequentially from the seed nanoparticle synthesis. All synthesis and regrowth processes were performed at constant stirring speed (900 rpm) and temperature (70 °C). Deionized water (Millipore-Q water, 18.2 MΩ cm) was used as solvent for all reactions.
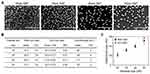
The size measurements of SNPs were performed by using SEM (FE-SEM; HITACHI S-4800) and DLS (ELS-Z, Otsuka electronics Co. Ltd.) analyses. In the SEM size analysis, the mean size and its distribution were derived using the data sets of measured ferret diameters in the SEM images of SNPs assuming that the size distribution follows log-normal function.15 We calculated the size and standard deviation of each nominal SNP based on evaluations of more than 100 particles in any region of the SEM image using Image-Pro Plus software. To obtain the DLS sizes (hydrodynamic diameter) of SNPs, 12 repetitive DLS measurements with 5 sub-runs were performed to give 12 cumulant size results. The six highest values of these 12 measurement results were then removed as a dust rejection filter in the calculation of the mean and polydispersity index (PDI) of the DLS size. The standard deviation of the size of the SNPs was calculated from the PDI values (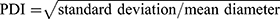 ). The surface charge (zeta potential) of SNPs was measured by using a laser Doppler velocimeter (Nanosizer Z, Malvern Instruments Ltd.). The mean and standard deviation of the zeta potential values were calculated from the results of 5 individual measurements with 20 repeated scans. The calibration for zeta potential measurement was done by using a transfer standard (DTS1050, Malvern Instruments Ltd.), which has a zeta potential of 50±5 mV.
). The surface charge (zeta potential) of SNPs was measured by using a laser Doppler velocimeter (Nanosizer Z, Malvern Instruments Ltd.). The mean and standard deviation of the zeta potential values were calculated from the results of 5 individual measurements with 20 repeated scans. The calibration for zeta potential measurement was done by using a transfer standard (DTS1050, Malvern Instruments Ltd.), which has a zeta potential of 50±5 mV.
Reagents and antibodies
Fluorescein isothiocyanate (FITC), (3-Aminopropyl) triethoxysilane (APTES), Chlorpromazine, Filipin III, and Amiloride were purchased from Sigma-Aldrich. Cell Proliferation Reagent WST‐1 was purchased from Roche Biochemicals. The primary antibodies used as follows. Anti-Cleaved Caspase-3 (Cell Signaling Technology), anti-Actin (Abclon), and anti-RIPK-1, anti-RIPK-3 (BD Bioscience).
Cytotoxicity assay
HepG2, A549, and SW480 cells were purchased from American Type Culture Collection (ATCC; HB-8065, CCL-185, and CCL-228) and cultured in DMEM (10% Fetal Bovine Serum (FBS), 1% Antibiotic-Antimycotic), and cells at passage two to three were used for all experiments. These cells were seeded at 1×104 cells/well in 96-well plates. After 24 h, cells were treated with SNPs of 20- to 50-nm with indicated concentrations (5, 10, 25, 50, 100, 200, 500 μg/mL) for 24 h. Then the medium was discarded carefully, 10% WST-1 solution was added into each well. After 2 h, cell viabilities was assessed by measuring absorbance at 450 nm on a microplate reader.
Cellular uptake
We prepared FITC-labeled 20-nm SNP to identify their cellular uptake. First, FITC-APTES conjugate solution was prepared by mixing 400 µl of butanol (Sigma Aldrich), 2 mg of FITC and 2 drops (using syringe) of APTES. The solution was stirred overnight avoiding light. The mixture of 400 µl of FITC-APTES solution, 12 mL of butanol and 18 mL of TEOS was added to 0.82 µM of L-arginine solution at 50°C. The temperature of the solution was kept at 50 °C for 18 h and then only aqueous phase was extracted by the using the separatory funnel. The FITC-labeled 50-nm SNP was synthesized in a manner similar to that described above. 2.1 mL of Ammonia solution (Sigma Aldrich), 5 mL of FITC-APTES conjugate solution and 0.75 mL of TEOS were added to 44.25 mL of ethanol in the 1 neck round-bottom flask. To wash the FITC- SNPs and change the solvent, centrifugation-wash-sonication process was repeated more than twice and then finally re-dispersed in deionized water.
HepG2 were seeded in 60 mm culture dishes and treated with FITC-SNPs in culture media containing 0.1% FBS. After incubated for 12 h at 37 °C under a 5% CO2 atmosphere, the medium was discarded carefully and washed out using 3% bovine serum albumin in phosphate-buffered saline (PBS). The cells were dissociated using Accutase (STEMCELL Technologies) for 10 min, fixed with 3% formaldehyde, and analysis by a flow cytometer (Beckton Dickenson FACS Calibur) to determine the cellular uptake of SNP. It was also confirmed by fluorescent microscopy. Cells were placed on 4-well chamber slides (1×104 cells/well) and treated with the FITC-SNPs as described above, and then fluorescence images were visualized using a Zeiss 510LSM META laser-scanning microscope (Carl Zeiss AG, Oberkochen, Germany). Endocytosis inhibitors were pretreated in the cells to determine whether SNPs were internalized by endocytosis. HepG2 cells were pre-incubated with Chlorpromazine (2 μg/mL), Filipin III (2 μg/mL), and Amiloride (50 μM), following to treatment of SNPs.
Apoptosis and necrosis analysis
Apoptosis and/or necrosis of the SNPs was determined by the FITC-Annexin-V and Propidium Iodide (PI) assay. HepG2 cells were seeded in 60 mm-plates for 24 h and SNPs were treated in low serum-containing condition. Then, cells were washed with PBS, harvested, and double-stained with FITC-Annexin-V and PI (BD bioscience), according to the manufacturer’s instructions. Cells were analyzed by flow cytometry using FACSCalibur (BD bioscience) and data analysis was performed using FlowJo software.
Western blot analysis and immunoprecipitation
For Western blotting of cleaved caspase-3 and RIPK3, HepG2 cells treated with SNPs were harvested under nondenaturing condition, remove media, and rinse with ice-cold PBS. Whole-cell protein extracts were resuspended by RIPA lysis buffer containing 50 mM Tris/Cl, pH 7.6, 150 mM NaCl, 1 mM EDTA, 0.5% Na-deoxycholate, 1% Triton X-100, 50 mM β-glycerophosphate, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 10 μg/mL Aprotinin, 10 μg/mL Leupeptin, and 0.5% NP-40. Cell lysates were resuspended 6× sample buffer, heated 95 °C and centrifuged. The samples were subjected to SDS-PAGE, and the proteins were transferred to polyvinylidene fluoride (PVDF, Millipore) membranes. The membranes were incubated with primary antibodies at 4 °C overnight. Then, the membranes were incubated with species-specific horseradish peroxidase (HRP)-conjugated secondary antibodies. The immunoreactive bands were visualized with a chemiluminescent substrate (GE Life Sciences).
For immunoprecipitation of RIPK1, cell lysates were prepared by NP-40 cell lysis buffer (50 mM Tris-Cl, 150 mM NaCl, 50 mM β-glycerophosphate, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 10 μg/mL Aprotinin, 10 μg/mL Leupeptin, and 1% NP-40). The cell lysates were pre-cleared with protein G-agarose beads (Millipore) at 4 °C for 1 h and incubated overnight at 4 °C with anti-RIPK-1 antibody. Next day, protein G-agarose was added and incubated 2 h. Then, the sample was washed with NP-40 lysis buffer and resuspended in 2× loading buffer, heated 95 °C and centrifuged.
Results
Preparation and characterization of SNPs
Well-defined SNPs of varying sizes were prepared using minor variations in the solvent-mixing ratio, as reported by Hartlen et al.14 The morphology and size of the SNPs were determined by analyzing SEM images. Spherical SNPs with nominal sizes of 20, 30, 40, and 50 nm had mean diameters of 22.4±4.3, 30.8±3.6, 40.2±4.4, and 51.4±7.0 nm, respectively (Figure 1A). As some SNPs can aggregate in an aqueous environment, the DLS sizes and zeta potentials of SNPs in phosphate-buffered saline (PBS) were determined using a Zetasizer. The PDI values of the SNPs of all sizes were measured to be less than 0.2, indicating a narrow size distribution (Figure 1B). The zeta potentials of the 20-, 30-, 40-, and 50-nm SNPs were determined to be −62±6, −38±6, −37±5, and −43±6 mV, respectively. No significant difference was found when comparing data from the SEM and DLS analyses (Figure 1C). It was previously reported that inadequate serum stability can potentially result in confusing results with respect to size-dependent cytotoxicity, due to SNP aggregation. Therefore, in this study, we decided to test SNPs under a low-serum condition (0.5%) for reliable assessment of size-dependent cytotoxicity.
Size-dependent effects of SNPs on various biological activities
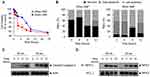
We evaluated various biological activities of SNPs according to size in three different human carcinoma cell lines (HepG2, A549, and SW480). First, the cell morphologies and viabilities were determined after treatment with various concentrations of SNPs (5–500 μg/mL) for 24 h (Figures 2A and S1). In all three types of cells, the cell viability decreased in a dose-dependent manner, and SNPs smaller than 30-nm in size were lethal to the cells. Further observations of cell morphology via optical microscopy revealed that these cells were significantly damaged. We performed a scratch “wound-healing” assay to assess the effect of SNPs on cell migration, because the indicated cell types exhibit some mobility in this type of assay (Figure S2). A linear scratch was introduced into a confluent monolayer of the cells, which were then cultured in serum-free medium to minimize cell proliferation. Microscopic observations to assess the capacities of the cells to fill the wound areas after treatments with SNPs with different sizes or doses showed that, in the absence of SNPs, substantial migration into the wound area occurred after 24 h. Treatment with SNPs less than 30-nm in size inhibited this migration in a dose-dependent manner, beginning at 50 μg/mL. In contrast, little significant inhibition of migration was detected after treatment with SNPs above 40-nm in size.
When exposed to SNPs, apoptotic cell death occurs in many cell types due to oxidative stress, such as elevated intracellular ROS levels.16 We determined intracellular ROS levels in the cells used in this study, which were treated with SNPs of various sizes for 24 h. Cellular ROS levels increased slightly in cells treated with SNPs of all sizes in a dose-dependent manner, and 20-nm SNP caused the largest increase in ROS levels (Figure S3). The 20- and 50-nm SNPs clearly displayed distinct biological activities. Many reports have demonstrated that both cytotoxicity and genotoxicity are closely associated with oxidative stresses.17,18 We investigated genotoxicity induced by 20- and 50-nm SNPs in these carcinomas cells. However, we did not observe significant genotoxicity differences based on the SNP size. At a relatively high concentration (100 μg/mL), some micronucleus formation was observed among the binucleated cells, in contrast to the predominantly dose-dependent ROS increase and cytotoxicity seen with SNPs. No significant micronucleus was observed in SW480 cells exposed to the SNPs (data not shown).
Induction of apoptosis and necrosis by SNPs in HepG2 cells
To clarify whether the 20- and 50-nm SNPs induced toxicity through different mechanisms, the same cytotoxic doses were first determined for each SNP size. The SNP concentrations that induced 50% inhibition of HepG2 cell growth (IC50 values) were calculated from the cell-viability curves. The IC50 values were 75.2, 82.6, 169.8, and 175.2 μg/mL for the 20-, 30-, 40-, and 50-nm SNPs, respectively. To determine whether cell death mediated by SNPs of different sizes at the IC50 resulted from apoptosis or necrosis, HepG2 cells were treated with 20- and 50-nm SNPs for 12 h, stained with fluorescein isothiocyanate (FITC)-conjugated annexin V and propidium iodide (PI), and subjected to flow cytometric analysis (Figure 2B). Unlike the expectation of an interesting difference, we found that both prominent apoptosis and necrosis were simultaneously induced with both SNPs. Furthermore, we found that both pro-caspase-3 cleavage and the interaction between kinase RIP1 (RIPK1) and kinase RIP3 (RIPK3) were substantially increased, indicating that SNPs induced apoptosis and necrosis, respectively (Figure 2C).
Dependence of time-lapse cytotoxic mechanisms on the SNP size
At predetermined time intervals, we then examined the viability curves of cells treated with 20- and 50-nm SNPs at the IC50 values. Although the cells eventually showed 50% survival after 24 h, distinct differences in the survival curves were found among the cells treated with SNPs of different sizes. While the survival of cells treated 50-nm SNPs decreased linearly over time, a rapid reduction in survival occurred within the first 3 h in cells treated with 20-nm SNP (Figure 3A). The cytotoxic mechanism of 20-nm SNP at after the initial exposure time (3 and 6 h) was confirmed by fluorescence-activated cell sorting (FACS) analysis, and necrosis induction was found to be dominant. In the case of the 50-nm SNP, the ratio of apoptosis induction was relatively high, but overall, both mechanisms were induced, with few difference in the ratio by exposure time (Figures 3B and S5). Treatment with 50-nm SNP time-dependently increased pro-caspase-3 cleavage and upregulated RIPK1/RIPK3 signaling (Figure 3C and D). These results demonstrate that apoptosis and necrosis were simultaneously involved in HepG2 cell death mediated by 50-nm SNP. Although the 20-nm SNP may have induced somewhat similar cytotoxic mechanisms, a remarkable RIPK1/RIPK3 interaction at the initial time was very interesting.
Using FACS analysis, the cell mask-aspect ratio versus the cell mask-region was plotted to first identify a population with a single-cell morphology. A gating region defining a normal cell population with an aspect ratio close to 1 was used for the experimental analysis. The excluded region (outside of the gated region) may have comprised a population of destroyed cells or cell debris. We observed that the gated cell population decreased significantly within 3 h after 20-nm SNP treatment (Figure S6). In contrast, cells treated with 50-nm SNPs were relatively abundant the gated area until 12 h post-treatment. Considering that nanoparticles reach the cytosol of cells by endocytosis and that it takes some time to induce participate in cytotoxicity, it can be assumed that the 20-nm SNP may have damaged cells in during the cell internalization process.
Differences in internalization behaviors with SNPs of different sizes
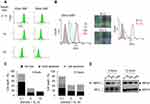
Next, we exposed cells to 20- and 50-nm SNPs at the same cytotoxic doses and closely examined their internalization behaviors. We prepared FITC-labeled SNPs and assessed their uptake in HepG2 cells via FACS analysis, based on the fluorescence intensities (Figure 4A). Based on the SEM and DLS analyses, we confirmed no significant difference in the size and distribution due to FITC-labeling (Figure S7). When the cells were treated with SNPs inducing the same level of cytotoxicity, similar SNP-internalization rates were observed. However, changes in the fluorescence intensities occurred over time, depending on the SNP size. Despite the short treatment time of 3 h, a considerable amount of 20-nm SNP was taken up into the cells, which was not significantly different from that found after 24 h (Figure S8). In contrast, internalization of the 50-nm SNP increased gradually over time (Figure S9). Consistent with the FACS analysis, each SNP showed two distinct internalization behaviors, as observed by fluorescent microscopy (Figure 4B). Most nanoparticles have been reported to enter into cells in a time-dependent manner, predominantly through endocytosis. Based on the data presented above, we hypothesized the 20-nm SNPs to enter cells by directly permeating the cell membrane, rather than through endocytic vesicles. To test this hypothesis, the cells were pretreated with an endocytosis inhibitor prior to treatment with each SNP, and our results showed that most of the 20-nm SNP were present in the cells, regardless of the inhibitor (Figures 4C and S10). In contrast, the endocytosis inhibitor significantly reduced intracellular uptake of the 50-nm SNP.
Effects of serum proteins on the cellular internalization of SNPs of different sizes
The serum stability of SNPs was tested in PBS solutions containing 1, 5, or 10% FBS. Figure 5A shows a low stability of aggregated SNPs following interaction with serum. In many reports, such instability was described for SNPs greater than 50-nm in the presence of serum, due to agglomeration.19 The agglomeration of SNPs may lead to somewhat confusing results with respect to size- or dose-dependent cytotoxicity. We treated HepG2 cells with 20-and 50-nm SNPs concentrations corresponding to their IC50 values in the presence of various serum concentrations (Figure 5B). The 50-nm SNPs, which increased to an average size >200 nm and showed excessive polydispersity in the presence of serum, did not affect cell survival after a 24 h exposure. Unlike the 50-nm SNP, the 20-nm SNP were relatively less affected by the serum and, in particular, monodispersity was maintained even at a 5% serum concentration. However, the average size inevitably increased to 50 nm due to agglomeration in serum, and the cytotoxicity was slightly reduced compared to those in the serum-free condition described above. Interestingly, the rapid reduction in surviving cells after the initial exposure time to 20-nm SNP was alleviated with increasing serum concentrations.
In an environment where the serum concentration exceeded 1%, the 50-nm SNP could barely enter into cells due to excessive aggregation between the particles and serum (Figure 6A). In contrast, the 20-nm SNP significantly entered the cells, even at a serum concentration of 5%. Surprisingly, the 20-nm SNPs exhibited a time-dependent internalization behavior that was completely different from that seen in a serum-free environment (Figure 6B). In addition, when the cells were pretreated with the endocytosis inhibitor, SNP internalization rarely observed, as opposed to the previous results showing that the 20-nm SNPs were not dependent on the endocytosis pathway (Figure S11). In a serum-free (0.1% serum) environment, approximately 60% of necrotic cell death occurred with only a 3 h exposure to the 20-nm SNP, whereas such cell death was dramatically reduced to around 10% after the SNPs had aggregated in the presence of serum (Figure 6C and Figure S12). In addition, RIPK1/RIPK3 signaling was down-regulated in 20-nm SNP-treated cells in the presence of serum, as confirmed by the Western blot shown in Figure 6D.
Discussion
Potential safety concerns related to biomedical and clinical applications of nanoparticles have been raised, necessitating the evaluation of their potential adverse effects on human health. Silica particles are deemed to be worthwhile as evaluation models to verify the potential risk of particles that are not biodegradable and accumulate in certain tissues. Moreover, silica particles of different sizes are used for various purposes in many research fields, including medicine, as they are one of the most easily exposed inorganic materials in human life. Since SNPs with a mesoporous structure can be easily loaded with specific cargoes such as drugs, they are the focus of more active research in applications as nanocarriers than any other type of inorganic nanoparticle. Hybrid platforms bearing other inorganic nanoparticles in these pores have recently been developed. Given this broad application potential, it is important to more closely assess the risks of SNPs.
An understanding of the causal relationship between the intrinsic biological properties of SNPs such as their sizes, shapes, agglomeration states, and surface-physicochemical properties, as well as their effects on human health, is critical for their safety in biomedical applications.20 However, the sizes of agglomerated SNPs and their underlying mechanisms of action and, thus, their toxicological effects remain poorly understood. Some evidence suggests that the cytotoxicity of SNPs is due to internalization via endocytosis, following exposure for a certain periods. Furthermore, it has been reported that SNPs approximately 100 nm in size are safe in a wide range of animals.21,22 The development of smaller SNP nanoparticles using the recent Stöber method, which is mainly used to synthesize SNPs (<100 nm in size), is expected to enable enhanced biological effects.23 Unfortunately, faster or stronger cytotoxicity is also likely to be induced, perhaps through a distinct pathway, depending on the size.
Several studies have shown that smaller nanoparticles are more cytotoxic, although these results are still controversial. Amorphous SNPs below 100 nm in size induced cytotoxicity in a size-dependent manner at 10–200 μg/mL doses.24 In their experiments, the authors examined the cytotoxicity of amorphous SNPs with average diameters of 30, 48, 118, and 535 nm in mouse keratinocyte cells. However, in another study where HepG2 cells were exposed to SNPs with sizes of 7, 20, and 50 nm at doses of 20–640 μg/mL, it was found that the cytotoxicity decreased in the order of 20>7>50 nm.25 Recently, in vitro toxicity was assessed in more detail using monodispersed spherical SNPs with sizes of 20, 40, 60, 80, 100, and 200 nm in A549, HepG2, and NIH/3T3 cells.26 The authors suggested that the SNP cytotoxicity was not only size- and dose-dependent, but was also cell-type dependent. To examine the size effects of SNPs on cytotoxicity, the most important prerequisite is the ability to control the experimental protocols, based on an understanding of the changes in physiological properties, along with the facile control of SNP sizes. Here, the complete control of serum agglomeration related to SNP-size variation was crucial for determining the biological responses to SNPs. We clearly demonstrates that the smaller the SNP size, the greater the toxicological activities in three cell lines. Of these, only the genotoxicity assessment did not show a significant dependence upon the SNP size or dose.
Even if smaller nanoparticles are evaluated in preclinical animal models, accurate validation of differences in their toxic effects will be difficult. Furthermore, the toxicological results may show comparable safety when compared to existing data with larger SNPs. Such an outcome could be due to SNP agglomeration with serum proteins, resulting in the formation of complexes hundreds of nanometers in size.27 Assessment of SNP toxicity when delivered to a target site in such an agglomerated state is insufficient for determining the potential toxicities of SNPs exposed to multiple tissues. Therefore, it should be examined whether SNPs induce tissue or cellular toxicity after penetrating throughout the tissue with subsequent exposure for a significant period of time. In particular, we speculate that size-dependent toxicity will occur without the effects of serum when SNPs accumulate in interstitial tissue fluids with little serum protein content. If so, prior to animal testing to assess the potential toxicities of SNPs according to size, the size-dependent cellular-internalization behavior of SNPs under agglomeration-free conditions and the associated mechanisms of toxicity should be studied. Furthermore, it is worth examining whether the toxic mechanisms change following serum aggregation. Here, we synthesized well-defined SNPs of different sizes to explore the resulting size-dependent cellular responses. Because nanoparticles are known to accumulate in the liver,28 our detailed toxicological evaluations were performed in HepG2 liver cells.
The 20-nm SNP, but not SNPs of other sizes, showed significant adverse effects on biological activities such as cell viability and mobility, which were due to cytotoxicity rather than genotoxicity (Figures S1–S4). In each cell type, cell viability and migration decreased, while ROS production increased, in dose- and size-dependent manners, with SNPs <30 nm showing the greatest effects. Morphological damage was also apparent after SNP exposure. However, significant size-dependent genotoxic differences were not observed. ROS-mediated oxidative stress is a typical cytotoxic effect of engineered nanoparticles such as SNPs.29 SNP-induced apoptosis mechanisms have recently been reported in various cell lines, verifying a critical role for ROS. Our results were consistent with remarkable ROS-induced cytotoxicity and were observed in a size-dependent manner. Both 20-nm and 50-nm SNPs induced apoptosis via the caspase-cascade pathway. Necroptosis is another key cause of cytotoxicity and is investigated by co-immunoprecipitation of RIPK1 and RIPK3. SNP treatment significantly increased RIPK3 activation by RIPK1, indicating that they also induced necrosis. We previously demonstrated that human endothelial cells exposed to the SNPs underwent both apoptosis and necroptosis by inducing ROS-mediated endoplasmic reticulum stress and autophagy, respectively.30 More importantly, this finding provides a reliable means for studying the size-dependent cytotoxicity of SNPs in HepG2 cells. To examine the size-dependent cytotoxic mechanisms of SNPs, size variation due to serum was minimized in serum-free medium, and the evaluation was performed based on the IC50 values.
Using 20-nm and 50-nm SNPs at the IC50, apoptosis and necrosis occurred simultaneously in SNP-treated HepG2 cells. After 12 h, unfortunately, it was unclear whether the difference in the cytotoxic mechanism depended on the SNP size (Figure 2B and C). It is probable that the difference in IC50 values observed over time reflected the internalization efficiency, not a specific cytotoxic mechanism. This possibility is supported by the observation that internalization of the 50-nm SNP increased over time (Figure 4A). Using the same SNP concentration, the smaller 20-nm SNP showed a stronger fluorescence intensity, indicative of greater cellular uptake (data not shown). Here, we assumed that different cytotoxic mechanisms would be observed according to the exposure time. Cells were exposed to SNPs of different sizes at their respective IC50 values, and cell viability was observed over time. Surprisingly, a drastic reduction in cell viability occurred within 3 h. As RIPK1/RIPK3 signaling was upregulated, these cells appeared to have initiated necrosis (Figure 3D). We propose that the cause of these different cytotoxic mechanisms depended on the route of intracellular uptake of the 20-nm SNP, which differed from that of the 50-nm SNP. Nanoparticles with sizes of tens to hundreds of nanometers can enter cells by endocytosis. In addition, many studies of size-dependent endocytosis of nanoparticles have been reported.31 These results provide a sufficient basis for concluding that the efficiency of endocytosis increases as the size becomes smaller. However, there is not support the interpretation that 20-nm SNPs are rapidly transferred to the cells, thereby leading to necrotic cell death. In particular, we clearly demonstrated that the 20-nm SNPs were internalized into cells, despite pretreatment with an endocytosis inhibitor (Figure 4C).
A previous study of the intracellular fate of SNPs <50 nm in size using transmission electron microscopy provides a plausible basis for early necrosis.32 Data from that study showed that the cell membranes were destroyed when SNPs below 20 nm were internalized. Although only morphological changes were investigated, the possibility of necrosis induction via membrane disintegration was fully considered, whereas no significant apoptosis occurred in the cells destroyed by the SNPs. Here, we addressed the mechanism of cytotoxicity in detail at the point of initial exposure, when the cell membranes would be impacted (Figure 3). Some Annexin-V-positive cells and caspase-3 activation were observed, independently of their sizes. However, a high proportion of PI-positive cells was observed after exposure to 20-nm SNP, and a prominent interaction between RIPK1 and RIPK3 was observed. Thus, SNPs of a certain size (<20 nm) may cause necrosis by damaging the cell membrane during internalization. Collectively, these results suggest that SNPs, when deeply distributed throughout various tissues and their constituent cells, may be more toxic than those evaluated in many studies if they are 20 nm in size.
Finally, we verified the effects of serum agglomeration on these internalization and cytotoxic mechanisms. The agglomerated 20-nm SNP, determined to be 50 nm in size following serum exposure, gradually entered the cells via endocytosis, as if they were originally 50-nm SNPs without agglomeration. Likewise, necrosis induction, which caused toxicity even after a short exposure, markedly decreased in cells treated with agglomerated SNPs. The agglomerated SNPs (simulating the effects of serum exposure) may have been unable to penetrate the cell membrane and induce necrosis; therefore, the contact of SNPs with the cells was relatively reduced. These results are in part supportive of our suggestion that the 20-nm SNP showed the unique internalization behavior that was closely related to early necrotic cell death. Whether this change in intracellular uptake was reflected the influence of the surrounding serum or the increased size cannot be deduced from our results. However, it is clear that 20-nm SNP caused strong necrosis despite a short exposure time, which should be reviewed for potential size-dependent toxicity with SNPs in a serum-poor environment.
Cytotoxicity and mechanistic studies of SNPs in serum-free environments indicated a unique and more lethal risk of 20-nm SNP. We examined the size-dependent effects of SNPs on apoptosis and necrosis with human hepatoma (HepG2) cells. Both apoptosis and necrosis occurred in serum-free conditions following exposure to 20-nm or 50-nm SNPs, and only cells treated with 20-nm SNPs exhibited relatively rapid necrosis. In contrast to the 50-nm SNPs, inhibiting endocytosis did not impede internalization of the 20-nm SNPs. Agglomeration following serum exposure increased the size of the 20-nm SNP to approximately 50 nm, and internalization and cell membrane damage were no longer observed; thus, early necrosis was inhibited. Our results suggest that safety guidelines should be revised to account for size-dependent cytotoxicity, considering that the 20-nm SNP clearly elicited stronger cytotoxic effects and early necrotic cell death in serum-free conditions, which may be similar to the microenvironment of SNPs after tissue penetration. In addition to SNPs, several other types of inorganic nanoparticles may or may not destroy cell membranes below a certain size; however, our toxicity studies are considered to be a worthwhile assessment for establishing safety guidelines for a given size.
Acknowledgment
The authors thank Drs. Sang Chul Lee, Baek-Soo Han, and Kyoung-Jin Oh for their helpful advice. This study was supported by a grant from the Korea Research Institute of Bioscience and Biotechnology and by grants from the National Research Foundation of Korea, which was funded by the Ministry of Science, Information & Communication Technology and Future Planning (NRF-2015M3A9C6030284, 2015M3A9D7029882, 2015M3A7B6027948).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Khan I, Saeed K, Khan I. Nanoparticles: properties, applications and toxicities. Arab J Chem. 2017. in press. doi:10.1016/j.arabjc.2017.05.011
2. Srivastava V, Gusain D, Sharma YC. Critical review on the toxicity of some widely used engineered nanoparticles. Ind Eng Chem Res. 2015;54:6209–6233. doi:10.1021/acs.iecr.5b01610
3. Katz LM, Dewan K, Bronaugh RL. Nanotechnology in cosmetics. Food Chem Tox. 2015;85:127–137. doi:10.1016/j.fct.2015.06.020
4. Peters RJ, Bouwmeester H, Gottardo S, et al. Nanomaterials for products and application in agriculture, feed and food. Trends Food Sci Technol. 2016;54:155–164. doi:10.1016/j.tifs.2016.06.008
5. Mishra A, Kumar J, Melo JS. An optical microplate biosensor for the detection of methyl parathion pesticide using a biohybrid of Sphingomonas sp. cells-silica nanoparticles. Biosen Bioelect. 2017;87:332–338. doi:10.1016/j.bios.2016.08.048
6. Wang Y, Zhao Q, Han N, et al. Mesoporous silica nanoparticles in drug delivery and biomedical applications. Nanomed. 2015;11:313–327. doi:10.1016/j.nano.2014.09.014
7. Liu J, Luo Z, Zhang J, et al. Hollow mesoporous silica nanoparticles facilitated drug delivery via cascade pH stimuli in tumor microenvironment for tumor therapy. Biomat. 2016;83:51–65. doi:10.1016/j.biomaterials.2016.01.008
8. Salatin S, Maleki Dizaj S, Yari Khosroushahi A. Effect of the surface modification, size, and shape on cellular uptake of nanoparticles. Cell Biol Int. 2015;39:881–890. doi:10.1002/cbin.10459
9. Izak-Nau E, Voetz M, Eiden S, et al. Altered characteristics of silica nanoparticles in bovine and human serum: the importance of nanomaterial characterization prior to its toxicological evaluation. Part Fibre Tox. 2013;10:56. doi:10.1186/1743-8977-10-56
10. Murugadoss S, Lison D, Godderis L, et al. Toxicology of silica nanoparticles: an update. Arch Tox. 2017;91:2967–3010. doi:10.1007/s00204-017-1993-y
11. Fröhlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int J Nanomed. 2002;7:5577–5591.
12. Oh N, Park J-H. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int J Nanomed. 2014;9:51–63.
13. Alkhammash HI, Li N, Berthier R, de Planque MR. Native silica nanoparticles are powerful membrane disruptors. Phys Chem Chem Phys. 2015;17:15547–15560. doi:10.1039/c4cp05882h
14. Hartlen KD, Athanasopoulos AP, Kitaev V. Facile preparation of highly monodisperse small silica spheres (15 to >200 nm) suitable for colloidal templating and formation of ordered arrays. Langmuir. 2008;24:1714–1720. doi:10.1021/la7025285
15. Song NW, Park KM, Lee I-H, Huh H. Uncertainty estimation of nanoparticle size distribution from a finite number of data obtained by microscopic analysis. Metrol. 2009;46:480. doi:10.1088/0026-1394/46/5/012
16. Ahmad J, Ahamed M, Akhtar MJ, et al. Apoptosis induction by silica nanoparticles mediated through reactive oxygen species in human liver cell line HepG2. Tox Appl Pharm. 2012;259:160–168. doi:10.1016/j.taap.2011.12.020
17. Hanot-Roy M, Tubeuf E, Guilbert A, et al. Oxidative stress pathways involved in cytotoxicity and genotoxicity of titanium dioxide (TiO2) nanoparticles on cells constitutive of alveolo-capillary barrier in vitro. Tox in Vitro. 2016;33:125–135. doi:10.1016/j.tiv.2016.01.013
18. Gallo A, Manfra L, Boni R, et al. Cytotoxicity and genotoxicity of CuO nanoparticles in sea urchin spermatozoa through oxidative stress. Env Int. 2018;118:325–333. doi:10.1016/j.envint.2018.05.034
19. Ahmad A, Zakaria N, Lockman Z, Razak K. The effect of silica nanoparticles stability in biological media. J Phys. 2018;1082:012047.
20. Gatoo MA, Naseem S, Arfat MY. et al. Physicochemical properties of nanomaterials: implication in associated toxic manifestations. Biomed Res Int. 2014. 1–8. doi:10.1155/2014/498420
21. Xie G, Sun J, Zhong G, et al. Biodistribution and toxicity of intravenously administered silica nanoparticles in mice. Arch Tox. 2010;84:183–190. doi:10.1007/s00204-009-0488-x
22. Argyo C, Weiss V, Bräuchle C, Bein T. Multifunctional mesoporous silica nanoparticles as a universal platform for drug delivery. Chem Mat. 2013;26:435–451. doi:10.1021/cm402592t
23. Meier M, Ungerer J, Klinge M, Nirschl H. Synthesis of nanometric silica particles via a modified Stöber synthesis route. Coll Surf A. 2018;538:559–564. doi:10.1016/j.colsurfa.2017.11.047
24. Yu Y, Li Y, Wang W, et al. Acute toxicity of amorphous silica nanoparticles in intravenously exposed ICR mice. PLoS One. 2013;8:e61346. doi:10.1371/journal.pone.0061346
25. Lu X, Qian J, Zhou H, et al. In vitro cytotoxicity and induction of apoptosis by silica nanoparticles in human HepG2 hepatoma cells. Int J Nanomed. 2011;6:1889–1901.
26. Kim I-Y, Joachim E, Choi H, Kim K. Toxicity of silica nanoparticles depends on size, dose, and cell type. Nanomed. 2015;11:1407–1416. doi:10.1016/j.nano.2015.03.004
27. Orts-Gil G, Natte K, Drescher D, et al. Characterisation of silica nanoparticles prior to in vitro studies: from primary particles to agglomerates. J Nanopart Res. 2011;13:1593–1604. doi:10.1007/s11051-010-9910-9
28. Zhang Y-N, Poon W, Tavares AJ, et al. Nanoparticle–liver interactions: cellular uptake and hepatobiliary elimination. J Cont Rel. 2016;240:332–348. doi:10.1016/j.jconrel.2016.01.020
29. Manke A, Wang L, Rojanasakul Y. Mechanisms of nanoparticle-induced oxidative stress and toxicity. Biomed Res Int. 2013;1–15. doi:10.1155/2013/942916
30. Lee K, Lee J, Kwak M, et al. Two distinct cellular pathways leading to endothelial cell cytotoxicity by silica nanoparticle size. J Nanobiotechnol. 2019;17:24. doi:10.1186/s12951-019-0456-4
31. Kou L, Sun J, Zhai Y, He Z. The endocytosis and intracellular fate of nanomedicines: implication for rational design. Asian J Pharm Sci. 2013;8:1–10. doi:10.1016/j.ajps.2013.07.001
32. Messerschmidt C, Hofmann D, Kroeger A, et al. On the pathway of cellular uptake: new insight into the interaction between the cell membrane and very small nanoparticles. Beilstein J Nanotechnol. 2016;7:1296–1311. doi:10.3762/bjnano.7.121
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.