Back to Journals » Infection and Drug Resistance » Volume 16
A Rare Strain Actinomadura geliboluensis Was First Isolated from the Bronchoalveolar Lavage Fluid of a Patient with Pneumonia
Authors Yu Y , Yang G, Wang Y , Jin F , Wang H , Yu Z , Li L, Li X, Gao J, Xu W
Received 23 February 2023
Accepted for publication 11 May 2023
Published 18 May 2023 Volume 2023:16 Pages 3101—3108
DOI https://doi.org/10.2147/IDR.S409701
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yefu Yu,1,* Guier Yang,2,* Yanan Wang,1 Faxiang Jin,1 Huiyu Wang,1 Zhongqiang Yu,3 Lanqing Li,4 Xiangcheng Li,4 Junshun Gao,4 Wenfang Xu1
1Department of Clinical Laboratory, Affiliated Hospital of Shaoxing University, Shaoxing, 312000, People’s Republic of China; 2Emergency Ward of Affiliated Hospital of Shaoxing University, Shaoxing, 312000, People’s Republic of China; 3Department of Imaging, Affiliated Hospital of Shaoxing University, Shaoxing, 312000, People’s Republic of China; 4Key Laboratory of Precision Medicine in Diagnosis and Monitoring Research of Zhejiang Province, Hangzhou, 310016, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wenfang Xu, Department of Clinical Laboratory, Affiliated Hospital of Shaoxing University, 999 Zhongxing South Road, Yuecheng District, Shaoxing, People’s Republic of China, Tel +8613857589470, Email [email protected] Junshun Gao, Key Laboratory of Precision Medicine in Diagnosis and Monitoring Research of Zhejiang Province, 3 Qingchun East Road, Shangcheng District, Hangzhou, People’s Republic of China, Email [email protected]
Background: Actinomadura geliboluensis was first isolated in 2012 in Gelibolu, Canakkale, Turkey, and has not been reported to be isolated from humans until now. We have isolated it from the bronchoalveolar lavage fluid (BLF) of a patient with pneumonia and found its drug resistance. It is the first time that Actinomadura geliboluensis has been isolated from humans since its discovery and naming. This case may provide new ideas and methods for the clinical diagnosis and treatment of pulmonary actinomycosis.
Case Description: The patient was a 75-year-old male who was hospitalized in a township hospital and failed to improve after penicillin treatment. After admission to our hospital, the patient was treated with piperacillin/tazobactam according to clinical guidelines for 14 days. Actinomadura geliboluensis was isolated from the patient’s BLF and was identified by 16S rRNA sequencing. This report shows the biological characteristics and in vitro drug susceptibility testing, as well as the genomics analysis based on next-generation sequencing (NGS). The results demonstrated that Actinomadura geliboluensis was easy to be mistakenly identified as Actinomyces dental caries by using the Merieux ANC identification card. Based on the MIC test, Actinomadura geliboluensis was susceptible to tetracyclines, quinolones and sulfonamides, but resistant to carbapenems, penicillins and cephalosporins. The K-B test results showed Actinomadura geliboluensis was highly sensitive to piperacillin/tazobactam. Genomic analysis based on NGS showed that the Actinomadura geliboluensis belongs to Planobispora rosea EF-Tu mutants conferring resistance to inhibitor GE2270A, AAC(3)-VIIa, vanRO, chrB, and mexY.
Conclusion: Actinomycetes is generally sensitive to Penicillin but Actinomadura geliboluensis is not. In vitro drug susceptibility test is needed to support individualized drug use to avoid delay in the disease.
Keywords: Actinomadura geliboluensis, pulmonary actinomycosis, antibiotic resistance, next generation sequencing, genomics
Introduction
Pneumonia is a common and serious respiratory disease. The problem of antibiotic resistance has plagued the clinical treatment of pneumonia. With the development of molecular biological detection methods, 16S rRNA sequencing and Next-generation sequencing (NGS) are widely used in clinical microbial detection and functional identification. The molecular biological analysis of clinically selected strains can accurately determine the species, and the related pathogenicity and drug resistance can be obtained, which has a certain guiding significance for clinical drug use.
Actinomadura geliboluensis belongs to Actinomadura. It was first isolated in 2012 in Gelibolu, Canakkale, Turkey, and has not been reported to be isolated from humans until now.1,2 We have analyzed a strain isolated from the bronchoalveolar lavage fluid (BLF) of a patient with pneumonia. The strain was Actinomadura geliboluensis successfully identified by 16S rRNA sequencing after failing to be identified by biochemical and mass spectrometry way. In this case, the clinical laboratory-based biological characteristics and drug susceptibility profile of this strain were studied, and the genomic analysis based on next-generation sequencing (NGS) was performed. This is the first report that Actinomadura geliboluensis was isolated and studied from the human body, which may provide new ideas and methods for the clinical diagnosis and treatment of pulmonary actinomycosis.
Case Presentation
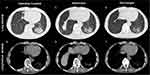
The patient, a male, aged 75 years, was admitted to our hospital (Affiliated hospital of Shaoxing University) with silicosis and community-acquired pneumonia following “cough, expectoration, and fever for 2 weeks”. The patient described himself as a farmer who worked part-time as a sandblaster during his free time. He had a 3-year history of dust exposure and a 40-year history of smoking, with an average of 20 cigarettes per day. Two weeks ago, the patient had a cold cough and sputum, which was paroxysmal and moderate in degree, with a small amount of white sputum and blood in the sputum, which was not easy to cough up, accompanied by fever. At that time, body temperature was not measured, and the patient had no fear of chills and shivering, no headache or dizziness, no fatigue, and night sweats. The patient was first hospitalized at a township health center. Some laboratory tests showed no abnormalities in liver function, renal function, blood lipids, myocardial enzymes, and electrolytes. Tests for antibodies to seven respiratory viruses were negative. Abnormal terms included WBC (3.9×10^9/L), N (80%), L (14%), CRP (38.4 mg/L). Computerized tomography (CT) examination showed interstitial changes in both lungs, left lower lung infection, and multiple mediastinal and hilar lymph nodes with calcification (Figure 1). The patient was treated with ampicillin sodium 4g bid+ iparfloxacin sulfate 0.4g qd for 10 days for infection, cough, and expectoration for about 10 days. The fever subsided but the patient still had persistent paroxysmal cough and expectoration. Finally, the patient came to our hospital for treatment.
A physical examination revealed that the temperature of 36.8°C, the pulse of 87 beats/min, the breath of 19 beats/min, and the blood pressure of 126/83 mmHG. The patient was conscious and spirited. The lips were not cyanosis, the jugular vein was not filling, the neck was soft, and the breath sounds of the lungs were thick. The patient had no obvious dry and wet rales, and with a normal heart rhythm. There was no obvious bruit, the abdomen was flat and soft, and the liver and spleen were not under the ribs. There was no tenderness and rebound pain, and the lower limbs were not having obvious edema. There were no fresh petechiae on the skin and no positive signs on neurological examination.
CT showed increased translucency of both lungs, scattered patchy and lamellar increased density shadows in both lungs and clump-like high-density shadows with unclear boundaries in the lower lobe of the left lung (Figure 1). There was no obvious displacement of the mediastinal position, and multiple mediastinal and bilateral hilar lymph nodes were enlarged with calcification. There were no obvious abnormalities in the shape and size of the great vessels in the heart and lungs. There was thickening and adhesion of pleura on both sides, no pleural effusion was found, and diffuse lesions of both lungs were observed. Silicosis with infection and tuberculosis were considered. Finally, the patient was admitted to the hospital with silicosis and community-acquired pneumonia.
After admission, relevant examinations were performed, and the 2019-nCoV nucleic acid test was negative. Blood gas analysis showed temperature 36.7°C, blood pH 7.403, partial pressure of carbon dioxide 41.8 mmHg, partial pressure of oxygen 87.9 mmHg, bicarbonate 1.1 mmol/L, standard bicarbonate 1.3 mmol/L, oxygen saturation 98%, lactic acid 1.7 mmol/L, Red blood cell count 3.76×1012/L, hemoglobin 120 g/L, white blood cell count 5.6×109/L, neutrophil classification 82%, lymphocytes 11%, monocytes 6%, reactive protein 2.2 mg/L, prothrombin time of 12.9 seconds, plasma D-dimer 670 μg/L. Plasma fibrinogen was 4.86 g/L. Liver function tests showed slight hypoproteinemia (except TP 57.2 g/L and albumin 38.7 g/L). Other serum biochemical and tumor tests were normal. The routine electrocardiogram showed sinus rhythm. Pulmonary function test showed mild restrictive ventilatory dysfunction and negative bronchodilation examination.
According to the Chinese Guidelines for the Diagnosis and Treatment of adult community-acquired pneumonia (2016 edition), the CURB-65 score was score 2. Considering the patient’s advanced age, pneumoconiosis, and recurrent infection, the patient was treated with piperacillin-tazobactam 3.375 g q8h by intravenous drip to cover gram-negative bacteria and positive bacteria, and ambroxol to relieve cough and expectoration and other symptomatic treatment, and the relevant examination. The treatment plan was adjusted at any time according to the condition. The timeline of medications administered for patients in our hospital as shown in Table 1.
 |
Table 1 The Timeline of Medications Administered for Patients in Our Hospital |
The next day, the sputum culture showed normal bacterial growth. There was no pathogenic Haemophilus and a few Candida albicans. Three days later, bronchoscopy treatment: bronchoscopy + lavage. Lavage fluid was sent for microbiological examination. A subsequent examination of the BLF was negative for acid-fast bacilli on fluorescent staining. Routine cytological examination showed light red color, slightly cloudy transparency, and nucleated cell count of 100×106/L, mainly neutrophils.
A few Gram-positive filament-like branching hyphae were seen on a direct smear. Mycobacterium tuberculosis (by GeneXpert assay) was negative. BLF was inoculated with Columbia blood plate medium, chocolate medium, and Cormagar fungal chromogenic medium, the first two were cultured for 24 hours, and fungal cultures for 3 days showed no bacterial growth. However, a rare Actinomyces Madura named Actinomadura geliboluensis was cultivated during the culture of BLF Mycobacterium. The strain was found on the 6th day of the whole MGIT liquid mycobacterial culture and showed no obvious growth for 1–2 days. On the 3rd day, small sand-like colonies were observed in the Columbia blood plate medium, and on the 4th day, colonies with a diameter of 1–1.5 mm were observed. It is noteworthy that the colony emits an extremely strong sewer-like odor. The colony morphology is shown in the ultra micrography (Figure 2A). The colonies were hard to emulsify in saline, and thin filament-like mycobacteria could be seen by Gram staining after rubbing on the glass slide. The bacteria were soft and not easy to break, and the morphology was similar to that seen by BLF direct staining. Acid-fast staining and modified weakly acid-fast staining were negative (Figure 2B). The other two media continually cultivated for 7 days without growth. The original liquid culture tubes were kept at 37°C for several days, and flocculent growth and radial arrangement of the filament-like mycobacteria were observed on smear staining (Figure 2C). By 42 days, no M. tuberculosis or nontuberculous mycobacteria were found.
According to the program file of MALDI TOF, the direct method and the formic acid extraction method were used to identify the mass spectrometer, and the obvious peak shape was observed, but the identification results were unreliable. The identification positions suggested semi-green and semi-red, and the protein fingerprints are shown in (Figure 3).
 |
Figure 3 Protein fingerprints detected by MALDI TOF. |
Biochemical tests were performed to identify the strain. The results showed that catalase was strongly positive and oxidase was weakly positive. An Anaerobic and Corynebacteria (ANC) identification card (Merieux, France) was used for detection on a VITEK2 bacterial identification instrument. Odontolyticus was identified as Actino. odontolyticus with a credibility of 97% and a Bionumber of 2320000000005. The biochemical results of ANC card were: 05LeuA leucine arylamidase +, 07PheA phenylalanine arylamidase +, 08ProA L-proline arylamidase +, 13TyrA tyrosine arylamidase +, and the rest were negative.
However, subsequent 16S rRNA sequencing confirmed that the identification result was unreliable. 16S rRNA gene amplification showed a single clear fragment. The sequencing results were compared by BLAST, and the sequence similarity with other strains was as follows: Actinomadura geliboluensis (GenBank No.NR_109059.1) 100%; Actinomadura formosensis (GenBank No.NR_114410.1) 99.11%; Actinomadura meyerae (GenBank No.NR_029090.1) 98.86%.
To further verify this result, the strain was identified as Actinomadura geliboluensis based on the valid data of NGS. Virulence factors (VF) and antimicrobial resistance (AMR) related genes were analyzed by gene function analysis. The nucleic acid sequence of the protein-coding gene was compared with the VFDB database, and no virulence factor was found. However, the genomic analysis based on NGS showed that the Actinomadura geliboluensis belongs to Planobispora rosea EF-Tu mutants conferring resistance to inhibitor GE2270A, AAC(3)-VIIa, vanRO, chrB, and mexY.
The drug resistance of the strain was analyzed by in vitro drug sensitivity test through minimum inhibitory concentration (MIC) test based on the broth microdilution method and drug susceptibility disk diffusion method (K-B test), the results as shown in Table 2 and Table 3. The results showed that Actinomadura geliboluensis was susceptible to tetracyclines, quinolones and sulfonamides, but resistant to carbapenems, penicillins and cephalosporins. The patient’s piperacillin/tazobactam inhibition zone was large as 46 mm by K-B disk method. In addition, amikacin, minocycline, ciprofloxacin and levofloxacin all had an inhibition zone of more than 40 mm, while penicillin, oxacillin and cefoxitin had no inhibition zone, ceftazidime, cefotaxime and amoxicillin/clavulanic acid had an inhibition zone of 12–17 mm. Although K-B method is not the recommended method for susceptibility testing of aerobic actinomycetes, the results of K-B method are consistent with MIC method.
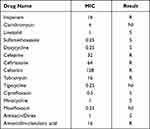 |
Table 2 Results of Drug Resistance Phenotypes of Actinomadura geliboluensis by MIC Drug Susceptibility Testing |
 |
Table 3 Results of Drug Resistance Phenotypes of Actinomadura geliboluensis by K-B Test |
After 14 days of treatment, the patient improved and was discharged. After 6 months of follow-up, the patient felt well and had no recurrence. The timeline of patient diagnosis and treatment has been shown in Figure 4.
 |
Figure 4 The timeline shows the entire diagnosis and treatment process of this case. |
Discussion
Actinomadura is a genus of bacteria under the family Thermomonosporaceae.3 It was once considered to be filamentous fungi but was later identified as filamentous bacteria by cell ultrastructural analysis and classified as aerobic actinomycetes.4 Among them were Actinomadura madurae, Actinomadura pelletieri, Actinomadura latina and A.palatopharyngis are associated with human disease.5–11 “Actinomadura geliboluensis” + “pulmonary actinomycosis” or “patient” or “human” were used as the search terms, and the search language was Chinese or English. No reports of Actinomadura geliboluensis in humans were retrieved from CNKI, Wanfang, and PubMed databases. This case first suggests that Actinomadura geliboluensis may also be associated with human disease. The DNA (G+C) content of Actinomadura was 66%~72%, and the model strain was Actinomadura madurae. The DNA (G+C) content of the case strain was 72.42% at a high value.
In the past, chest actinomycosis was usually characterized by mycetoma, with extensive destruction of the lung parenchyma and the chest wall.12,13 This case may be in the early stage, only showing pulmonary shadows on chest CT, and the infection can be treated with sensitive antibiotics to quickly improve the symptoms, which may be related to the early non-invasive infection. However, it is noteworthy that this strain may be resistant to macrolide antibiotics clarithromycin, aminoglycoside antibiotics tobramycin, penicillin antibiotics, cephalosporins antibiotics, and carbapenems antibiotics because of the presence of antibiotic resistance genes related to AAC (3)-VIIa, vanRO, chrB, and mexY. The identification of Actinomycetes of dental caries with the Biomerieux ANC card on the VETIK-2 bacterial identification instrument was easy to be mistakenly identified as high coincidence Actinomycetes of dental caries. It may cause clinicians to use penicillins empirically, resulting in the delay of the disease.
Piperacillin/tazobactam was suitable for the treatment of Actinomyces SPP. diseases, which was also verified by our drug susceptibility test. In addition, there was no growth of mycobacterium tuberculosis and nontuberculous mycobacteria in the routine bacterial culture of sputum and BLF. Only Actinomadura geliboluensis grew in BLF, which was sensitive to piperacillin/tazobactam and improved after clinical treatment.
Therefore, we believe that penicillin is considered to be the first drug for the treatment of actinomycosis and is not necessarily applicable to the treatment of Actinomadura geliboluensis. Tetracyclines, quinolones, and sulfonamide-sensitive antibiotics are recommended for empirical use. We hope to deepen the understanding of this bacterium and provide reference for the diagnosis and treatment of this bacterium relevant diseases in the future.
Conclusion
In conclusion, drug susceptibility testing is needed to support individualized drug treatment to avoid delays in the treatment of the disease. If the laboratory does not have the condition of MIC drug susceptibility test, the K-B method can be used to predict the results of drug susceptibility first, so as to provide the basis for clinical treatment of antibiotics as soon as possible.
Ethics Approval and Informed Consent
The authors certify that the patient consent form has been obtained. Written informed consent was obtained from the patient for publication of this Case report and any accompanying images. All procedures performed in the study involving human participants were in accordance with the ethical standards of the Ethics Committee of the Affiliated Hospital of Shaoxing University (Shaoxing Municipal Hospital). The ethics committee approved the waiver in this case report, based on the ethical standards to publish the case details.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; have drafted, revised or critically reviewed the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Medical Science and Health Science Program of Zhejiang Province (2020KY334).
Disclosure
Yefu Yu and Guier Yang contributed equally to this work and should be considered co-first authors. All authors report no conflicts of interest in this work.
References
1. Izri A, Aljundi M, Billard-Pomares T, et al. Molecular identification of Actinomadura madurae isolated from a patient originally from Algeria; observations from a case report. BMC Infect Dis. 2020;20:829. doi:10.1186/s12879-020-05552-z
2. Sazak A, Camas M, Spröer C, Klenk HP, Sahin N. Actinomadura geliboluensis sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2012;62:2011–2017. doi:10.1099/ijs.0.036145-0
3. Bonifaz A, Tirado Sánchez A, Vázquez González D, Fierro Arias L, Araiza J, González GM. Actinomycetoma by Actinomadura madurae. Clinical and therapeutic characteristics of 18 cases with two treatment modalities. J Dermatol Treat. 2020;33:954–958. doi:10.1080/09546634.2020.1793887
4. Bonifaz A, Tirado-Sánchez A, Vazquez-Gonzalez D, Araiza J, Hernández-Castro R. Actinomycetoma by actinomadura madurae: clinical characteristics and treatment of 47 cases. Indian Dermatol Online J. 2021;12:285–289. doi:10.4103/idoj.IDOJ_474_20
5. Ara I, Matsumoto A, Bakir MA, Kudo T, Ōmura S, Takahashi Y. Actinomadura bangladeshensis sp. nov. and Actinomadura chokoriensis sp. nov. Int J Syst Evol Microbiol. 2008;58(7):1653–1659. doi:10.1099/ijs.0.65533-0
6. Bakheet OE, Hassan MA, Fahal AH. Extensive perineal Actinomadura pelletieri actinomycetoma-induced urethral stricture: a rare complication. Trans R Soc Trop Med Hyg. 2021;115:415–419. doi:10.1093/trstmh/traa166
7. Bedi TR, Kaur S, Kumar B. Red grain mycetoma of the scalp (Actinomadura pelletieri). A case report from India. Mycopathologia. 1978;63:127–128. doi:10.1007/BF00441259
8. Cascio A, Mandraffino G, Cinquegrani M, et al. Actinomadura pelletieri mycetoma--an atypical case with spine and abdominal wall involvement. J Med Microbiol. 2011;60:673–676. doi:10.1099/jmm.0.027862-0
9. Ezaldeen EA, Ahmed RM, Wadella ES, Dawi NIE, Fahal AH. Cervical spinal cord compression: a rare and serious complication of Actinomadura pelletieri actinomycetoma. JMM Case Reports. 2015;2:14. doi:10.1099/jmmcr.0.000074
10. Siddig EE, Nyuykonge B, Ahmed M, et al. Human actinomycetoma caused by Actinomadura mexicana in Sudan: the first report. Trans R Soc Trop Med Hyg. 2020;115:406–410. doi:10.1093/trstmh/traa145
11. Trujillo ME, Goodfellow M. Polyphasic taxonomic study of clinically significant actinomadurae including the description of Actinomadura latina sp.nov. Zentralblatt fur Bakteriologie. 1997;285:212–233. doi:10.1016/S0934-8840(97)80029-1
12. Wu C, Huang SC, Qian S, et al. A case of pulmonary actinomycosis with protruding performance ipsilateral chest wall huge abscess. J Clin Infect Dis. 2019;12:12.
13. Łyżwa E, Siemion-Szcześniak I, Sobiecka M, et al. Pulmonary actinomycosis complicated by fistula of the chest wall. Adv Respir Med. 2021;89:532–537. doi:10.5603/ARM.a2021.0071
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.