Back to Journals » OncoTargets and Therapy » Volume 13
A Novel Serum Exosomes-Based Biomarker hsa_circ_0002130 Facilitates Osimertinib-Resistance in Non-Small Cell Lung Cancer by Sponging miR-498
Received 20 December 2019
Accepted for publication 15 May 2020
Published 9 June 2020 Volume 2020:13 Pages 5293—5307
DOI https://doi.org/10.2147/OTT.S243214
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Jing Ma,* Guanbin Qi,* Lei Li
Department of Respiratory and Critical Medicine, Huaihe Hospital of Henan University, Kaifeng, Henan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lei Li
Department of Respiratory and Critical Medicine, Huaihe Hospital of Henan University, No. 115 Ximen Avenue, Kaifeng, Henan 475000, People’s Republic of China
Tel +86-0371-23906701
Email [email protected]
Purpose: Exosomes are the effective delivery system for biological compounds, including circular RNAs. In this research, we aimed to explore the role of circular RNA hsa_circRNA_0002130 in osimertinib-resistant non-small cell lung cancer (NSCLC).
Materials and Methods: In our study, the relative protein expression of glucose transporter 1 (GLUT1), hexokinase-2 (HK2) and lactate dehydrogenase A (LDHA) was detected by Western blot, while the expression of hsa_circ_0002130 and microRNA-498 (miR-498) was detected by quantitative real-time PCR (qRT-PCR). The biological functions of hsa_circ_0002130 in osimertinib-resistant NSCLC were analyzed by cell viability assay, flow cytometry analysis, luciferase reporter assay, RNA pull-down assay, and tumor xenograft model in vivo. Moreover, glucose uptake, lactate production and extracellular acidification (ECAR) levels were measured by glucose uptake colorimetric assay kit, lactate assay kit II, and Seahorse Extracellular Flux Analyzer XF96 assay, respectively. hsa_circ_0002130 identification and localization were confirmed by RNase R digestion and subcellular localization assay, respectively. Exosomes were isolated from the sera collected from NSCLC patients and identified using a transmission electron microscopy and nanoparticle tracking analysis.
Results: Osimertinib-resistance was closely related to glycolysis. hsa_circ_0002130 was highly expressed in osimertinib-resistant NSCLC cells and hsa_circ_0002130 deletion inhibited osimertinib-resistance both in vitro and in vivo. Moreover, hsa_circ_0002130 targeted miR-498 to regulate GLUT1, HK2 and LDHA. The inhibitory effects of hsa_circ_0002130 deletion on osimertinib-resistant were reversed by downregulating miR-498. Importantly, hsa_circ_0002130 was upregulated in serum exosomes from osimertinib-resistant NSCLC patients.
Conclusion: Our findings confirmed that hsa_circ_0002130 served as a promotion role in osimertinib-resistant NSCLC.
Keywords: non-small cell lung cancer, hsa_circ_0002130, miR-498, osimertinib, exosome, GLUT1, HK2, LDHA
Introduction
Lung cancer is one of the most common human cancer with a high mortality rate in worldwide, and the incidence and death cases are 1.8 million new cases and 1.6 million death cases, respectively.1,2 Non-small cell lung cancer (NSCLC) is a type of lung cancer accounting for about 85%.3 Despite the therapeutic methods had achieved continuous improvements, the relapse and mortality were still the severe problems for NSCLC patients. Moreover, due to the delay in NSCLC diagnosis, the 5-year survival rate is extremely low that remains about 10%-15%.4 Currently, chemotherapy, radiotherapy, surgical excision, biological immunotherapy, and targeted molecular therapy were the main therapies for NSCLC patients, and EGFR-targeted therapy among these therapies for NSCLC gained wide attention worldwide.5,6 Osimertinib, a third-generation EGFR-tyrosine kinase inhibitor, has an effective prolongation for the survival of NSCLC patients at the advanced stage.7 However, the resistance to osimertinib in advanced NSCLC patients is inevitable. Thus, the exploration of the more molecular targets for osimertinib-resistant NSCLC patients is meaningful.
Circular RNAs (CircRNAs), a novel group of endogenous, abundant, and conserved non-coding RNAs, are characterized by a circular structure without 3ʹ poly (A) tails and 5ʹ caps.8,9 The covalently closed loop structure of circular RNA is generated through “back-splicing” with a downstream splice donor connected to an upstream splice acceptor, which made circRNAs more stable and resistant to the degradation of exonucleases than their linear counterparts.10 Growing evidence indicated that circRNAs exerted crucial effects in multiple biological processes, including cell survival, proliferation, differentiation, metastasis and apoptosis.11,12 CircRNAs were confirmed to combine with microRNAs (miRNAs) and served as sponges for miRNAs to regulate gene expression via a circRNA/miRNA/messenger RNA (mRNA) axis in human cancers, including NSCLC.13,14 For example, Han and his colleagues demonstrated that the oncogene circ-RAD23B could facilitate NSCLC progression, including cell growth and metastasis, through sponging miR-653-5p and miR-593-3p to regulate TIMA1 and CCND2, respectively.15 A novel circular RNA hsa_circ_0002130 upregulation was observed in osimertinib-resistant NSCLC cells.16 However, the potential mechanisms and functional effects of hsa_circ_0002130 in NSCLC remain unclear.
The aerobic glycolysis (Warburg effect), which could enhance tumor cell growth, metastasis and progression with facilitated lactate production and glucose uptake, is a feature of human cancer cells.17 Glucose transporter 1 (GLUT1) is a regulator in the increasing glucose uptake in glycolysis, which was revealed as a treatment target and diagnostic marker for various human cancers.18 Hexokinase 2 (HK2) is another critical driver of glycolysis, which is an enzyme that has the ability to convert glucose to glucose-6-phosphate participating in the rate-limiting step in glycolysis.19,20 Lactate dehydrogenase A (LDHA), which can catalyze pyruvate into lactate, was involved in the final step in glycolysis and confirmed to be associated with the prognosis of NSCLC patients.21 The detail mechanisms of the regulators, including GLUT1, HK2 and LDHA, are largely unknown, and the role of glycolysis in osimertinib-resistant NSCLC needs further investigated.
Exosomes are small intraluminal vesicles that generated and secreted by various types of cells with a diameter of about 30–150 nm, which can deliver bioactive cargoes, such as proteins, lipids, nucleic acids, circRNAs, long noncoding RNAs, miRNAs, mRNAs.22,23 Accumulating evidence suggested that exosomes served as vital mediators in cell-to-cell communication and exhibited critical effects on tumor growth and progression.24 Multiple studies demonstrated that exosomes could be secreted from cancer cells, and circRNAs were observed to be enriched in exosomes that might be diagnosis markers for cancers.25 However, little is known about the functional roles of secreted circRNAs in osimertinib-resistant NSCLC.
In the present paper, we analyzed the expression of hsa_circ_0002130 in osimertinib-resistant NSCLC cells and exosomal hsa_circ_0002130 expression in osimertinib-resistant NSCLC patients. Moreover, we explored the effects of hsa_circ_0002130 on tumor growth and metastasis. Mechanistically, we further investigated the potential mechanisms of hsa_circ_0002130 in osimertinib-resistant NSCLC progression.
Materials and Methods
Tissue Samples
Tissue samples were isolated from 28 osimertinib-resistant NSCLC patients (non-response) and 32 osimertinib-sensitive NSCLC patients (response), who were underwent surgery at Huaihe Hospital of Henan University. Written informed consents were obtained from all participates. This research was approved by the Human Research Ethics Committee of Huaihe Hospital of Henan University.
Cell Lines and Cell Culture
The two lung adenocarcinoma cell lines HCC827 and H1975 purchased from American Type Culture Collection (Manassas, VA, USA) were cultured in Roswell Park Memorial Institute 1640 (RPMI-1640) medium (Gibco, Carlsbad, CA, USA) supplementing with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA). 100 μg/mL streptomycin and penicillin (Invitrogen) were prior to adding into the RPMI-1640 medium to culture the cells. All cells were cultured in an incubator with the standard culture condition of 5% CO2 at 37°C. Osimertinib (AstraZeneca, Milan, Italy) was dissolved in dimethyl sulfoxide, followed by adding into the cell culture medium. Osimertinib-resistant HCC827 and H1975 cells (HCC827/OCR and H1975/OCR cell lines) were established as previously described using a dose-escalation method.16
Cell Transfection
Short hairpin RNAs (shRNAs) targeting hsa_circ_0002130 (sh-circ #1, sh-circ #2, sh-circ #3) and the control sh-NC, microRNA-498 (miR-498) mimic and the negative control (miR-NC), miR-498 inhibitor and NC inhibitor bought from Genepharma (Shanghai, China) were transfected into HCC827 and H1975 cells, as well as HCC827/OCR and H1975/OCR cells using Lipofectamine 3000 Reagent (Invitrogen).
Calculation of Half-Maximal Inhibitory Concentration (IC50)
HCC827, H1975, HCC827/OTR and H1975/OTR cells with a density of 1 × 104 cells per well were seeded into the 96-well plates. Then, the different concentrations of osimertinib including 0.0001 μM, 0.001 μM, 0.01 μM, 0.1 μM, 1 μM, 10 μM were added, respectively. After the treatment of osimertinib for 48 h, the absorbance of the cells on each well was detected at 450 nm by cell counting kit-8 (CCK-8) assay. IC50 represents the concentration of a drug that is required for 50% inhibition of cell growth, and IC50 was calculated using SPSS 18.0.
Determination of Glucose Uptake and Lactate Production
The level of glucose uptake was determined by glucose uptake colorimetric assay kit (BioVision, Milpitas, CA, USA). Briefly, cells with a density of 1 × 104 cells were seeded, and subsequently cultured with 100 mL Krebs-Ringer-Phosphate-HEPES (KRPH) buffer containing 2% albumin from bovine serum, followed by adding 10 mM 2-deoxyglucose (2-DG). The samples were collected for the analysis of the glucose uptake level. For the determination of lactate production, cells were seeded into the 96-well plate and starved using non-serum medium for 24 h. Then, the cells were suspended in the buffer from lactate assay kit II (BioVision). The glucose uptake colorimetric assay kit and lactate assay kit II (BioVision) were also used for the determination of glucose uptake and lactate production in tumor tissues from the nude mice according to the manufacturer’s instructions.
Measurement of Extracellular Acidification (ECAR)
The level of ECAR was determined by the Seahorse Extracellular Flux Analyzer XF96 (Seahorse Bioscience, North Billerica, MA, USA) in NSCLC resistant and sensitive cells. 2 × 104 NSCLC cells were cultured in the XF96-well plate overnight. Then, the cultured NSCLC cells were sequentially cultured with glucose, oligomycin (oxidative phosphorylation inhibitor), and 2-DG (glycolytic inhibitor). Seahorse XF-96 Wave software was used to analyze the data and ECAR detection was represented as mpH/min.
Western Blot Analysis
Total protein was isolated using RIPA lysis buffer (Beyotime, Shanghai, China). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate the proteins. Then, the proteins were incubated with the primary antibodies against GLUT1 (1:5000, ab40084, Abcam, Cambridge, UK), HK2 (1:1000, ab104836, Abcam), LDHA (1:2000, ab101562, Abcam), CD81 (1:1000, 10,037, Cell Signaling Technology, Boston, MA, USA), TSG101 (1:1000, ab30871, Abcam), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH,1:1000, ab8245, Abcam) overnight at 4°C. Then, the proteins were incubated with the secondary antibodies against goat anti-rabbit IgG H&L (1:2000, ab6721, Abcam) or goat anti-mouse IgG H&L (1:2000, ab205719, Abcam). Finally, the signals were visualized using an electrochemiluminescent system (PerkinElmer Life Science, Waltham, MA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from NSCLC cells or exosomes using Trizol reagent (Invitrogen). Briefly, total RNA was reverse-transcribed into complementary DNA using SuperScript III RT (Invitrogen), and then qRT-PCR was performed to determine the amplified transcript levels of the relative specific genes. U6 and GAPDH was used as the internal loads to normalize the expression of miR-498 and hsa_circ_0002130, respectively. The primer sequences of the relative genes were as follows, hsa_circ_0002130: CCACGTGGGAGATTCTGG (sense) and ACGTTCCACAGCCAGCTC (antisense), miR-498: TTTCAAGCCAGGGGGCGTTTTTC (sense) and GCTTCAAGCTCTGGAGGTGCTTTTC (antisense), U6: CTCGCTTCGGCAGCACA (sense) and AACGCTTCACGAATTTGCGT (antisense), GAPDH: AAGGCTGAGAATGGGAAAC (sense) and TTCAGGGACTTGTCATACTTC (antisense). The relative expression of miR-498 and hsa_circ_0002130 was measured by the 2 −∆∆Ct method.
RNase R Digestion
Total RNA isolated from HCC827/OTR and H1975/OTR cells were incubated with 6 units of RNase R (Epicenter Biotechnologies, Shanghai, China). After total RNA and RNase R incubation for 15 min at 37°C, the expression of hsa_circ_0002130 was examined using qRT-PCR.
Subcellular Localization
Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek Corp., Belmont, MA, USA) was used to measure the localization of hsa_circ_0002130. Firstly, the Lysis Buffer J was used to lyse the cells, and then cell lysates were centrifugated. Subsequently, the nuclear RNA and cytoplasmic RNA were added into anhydrous ethanol and Buffer SK, respectively. The cytoplasmic RNA and nuclear RNA were eluted by the spin column. Finally, the proportion of hsa_circ_0002130 in cytoplasmic and nucleus fractions was determined using qRT-PCR.
Cell Viability
The proliferation abilities of HCC827, H1975, HCC827/OTR and H1975/OTR cells were determined by CCK-8 assay. Briefly, the cells were plated into the 96-well plates and incubated for 24 h. Then, 10 μL CCK-8 solution (Dojindo, Tokyo, Japan) was added and the absorbance was measured at 450 nm.
Cell Apoptosis
Cell apoptosis was measured by flow cytometry analysis using an annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (BD Pharmingen, San Diego, CA, USA). After transfection for 48 h, cells were collected and washed. Then, cells were double-stained with 5 μL Annexin V- FITC and 5 μL propidium iodide (PI) in the dark for 15 min. Cell apoptotic ability was detected by a FACScan flow cytometer (Becton Dickinson, Mountain View, CA, USA).
Tumor Model
The 6–8 weeks female BALB/c nude mice were bought from Beijing Vital River Laboratory Animal Technology (Beijing, China). HCC827/OTR cells were stably transfected with sh-circ #1 or sh-NC. Approximately 2 × 106 sh- circ #1/sh-NC-infected HCC827/OTR cells were subcutaneously injected into the right flank of the nude mice. After 6 days, the mice were treated with osimertinib (5.0 mg/kg/d) by oral administration. All the mice were classified into three groups (5 per group): sh-NC group, sh-NC + osimertinib group, and sh-circ #1 + osimertinib group. The tumor volume was detected from 6 day every 3 days using a caliper and the tumor weight was detected after the mice euthanasia at 27 day. All the animal studies were performed in line with the Guide for the Care and Use of Laboratory Animals and approved by Huaihe Hospital of Henan University Experimental Animal Ethics Committee.
Dual-Luciferase Reporter Assay
The wide-type sequences of hsa_circ_0002130 (WT-hsa_circ_0002130), GLUT1 (WT-GLUT1 3ʹUTR), HK2 (WT-HK2 3ʹUTR) and LDHA (WT-LDHA 3ʹUTR) were constructed and inserted into pGL3 promoter vectors (Invitrogen). The mutant sequences of hsa_circ_0002130, GLUT1, HK2 and LDHA were used to establish hsa_circ_0002130 mutant (MUT-hsa_circ_0002130), GLUT1 mutant (MUT-GLUT1 3ʹUTR), HK2-mutant (MUT-HK2 3ʹUTR) and LDHA-mutant (MUT-LDHA 3ʹUTR) reporter vectors. HEK293T cells were co-transfected with WT-hsa_circ_0002130, WT-GLUT1 3ʹUTR, WT-HK2 3ʹUTR, WT-LDHA 3ʹUTR or MUT-hsa_circ_0002130, MUT-GLUT1 3ʹUTR, MUT-HK2 3ʹUTR, MUT-LDHA 3ʹUTR reporter vectors and miR-498 mimic or NC mimic. Finally, the relative luciferase activity was detected using a dual-luciferase reporter assay system (Promega, Madison, WI, USA).
RNA Pull-Down Assay
The biotin-coupled miR-498 probe and control oligo probe were synthesized by RiboBio (Guangzhou, China). Briefly, HCC827/OTR and H1975/OTR cells were lysed using lysis buffer. Cell lysates were incubated with the biotin-coupled miR-498 probe or biotin-coupled NC probe for 1 h at room temperature, and then the remaining cell lysates were cultured with streptavidin magnetic beads (Life Technologies, Mountain View, CA, USA). Subsequently, the lysis buffer was used to wash the beads. Finally, RNAs bound to the beads were extracted using TRIzol Reagent (Invitrogen), and then analyzed using qRT-PCR.
Exosome Isolation
Serum samples were collected from NSCLC patients who underwent surgical procedures at Huaihe Hospital of Henan University. All patients participating in the present research signed the written informed consent. The research was approved by the Human Research Ethics Committee of Huaihe Hospital of Henan University. Exosome isolation was essentially carried out as previously described.26 Briefly, the serum samples were irradiated for 48 h, and the medium was collected and centrifuged. The medium was centrifuged at 2000 × g for 10 min, and then 10,000 × g for 30 min for depleting cell debris. Then, exosomes were extracted using ultracentrifugation at 120,000 × g for 60 min. The pellet containing exosomes was washed using phosphate-buffered saline (PBS). Finally, the exosomes were suspended in PBS and stored at −80°C until quantitation. The Optima L-100XP ultracentrifuge (Beckman Coulter, Brea, CA, USA) was used for the ultracentrifugation. And the Nanoparticle tracking analysis (NTA) was carried out using a Nanosight NS300 instrument (Malvern Instruments, Malvern, UK).
Transmission Electron Microscopy (TEM)
Briefly, the exosomes suspended in PBS were absorbed onto a Formvar/carbon film-coated TEM grid (Alliance Biosystems, Osaka, Japan). After drying at room temperature for 10 min, the grids stained using phosphotungstic acid solution (2%, pH = 7.0). Then, the stained grids were observed using a transmission electron microscope (TEM) (JEM-2100F, JEOL, Japan).
Statistical Analysis
Data were reported as the mean ± the standard deviations (SD) and analyzed by SPSS 18.0 software. The significance of difference between groups was evaluated using Student’s t-test or One-way ANOVA. Pearson correlation analysis was used to analyze the relationship between miR-498 and GLUT1, HK2 and LDHA. A value of P < 0.05 was regarded as a statistically significant difference.
Results
Glycolysis Was Enhanced in Osimertinib-Resistant NSCLC Cells
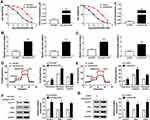
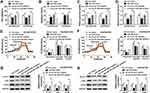
The osimertinib-resistant HCC827 cell line (HCC827/OTR) was established from the parental HCC827 cell line by gradually increasing the concentrations of osimertinib from 20.92 nM to 10 uM for six months. Meanwhile, H1975/OTR cell line was established from the parental H1975 cell line by gradually increasing the concentrations of osimertinib from 10.87 nM to 10 uM for six months. IC50 values of osimertinib for HCC827 and HCC827/OTR cells were 0.02092 uM and 1.278 uM, respectively. IC50 values of osimertinib for H1975 and H1975/OTR cells were 0.01087 uM and 0.5321 uM, respectively (Figure 1A). Subsequently, the glucose uptake and lactate production were detected in NSCLC sensitive and resistant cells. As shown in Figure 1B, the level of glucose uptake was significantly increased in HCC827/OTR and H1975/OTR cells compared with HCC827 and H1975 cells. Consistently, the level of lactate production was dramatically upregulated in HCC827/OTR and H1975/OTR cells relative to that in HCC827 and H1975 cells (Figure 1C). We also determined the ECAR level in NSCLC sensitive and resistant cells. We found an enhanced ECAR level in HCC827/OTR and H1975/OTR cells in comparison to HCC827 and H1975 cells (Figure 1D and E). Moreover, GLUT1, HK2 and LDHA were higher in HCC827/OTR and H1975/OTR cells than that in HCC827 and H1975 cells (Figure 1F and G). All these results indicated that the glycolysis was facilitated in osimertinib-resistant NSCLC cells.
hsa_circ_0002130 Was Upregulated in Osimertinib-Resistant NSCLC Cells
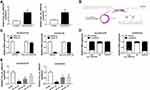
We discovered that hsa_circ_0002130 was increased in HCC827/OTR and H1975/OTR cells (Figure 2A). Moreover, we found that hsa_circ_0002130 was derived from the host gene C3 and consisted of 2 exons (exon 18–19), which was cyclized with the head-to-tail splicing of exon 18 and exon 19 according to circBase. The exist of back-splice junction was confirmed by our sanger sequencing (Figure 2B). Moreover, we performed RNase R digestion assay to verify the circular nature of hsa_circ_0002130. The results confirmed that hsa_circ_0002130 was indeed circRNA, which was resistant to RNase R digestion (Figure 2C). Subsequently, we measured the subcellular localization of hsa_circ_0002130 by nuclear and cytoplasmic separation experiments. The result suggested that hsa_circ_0002130 was mostly located in the cytoplasm of HCC827/OTR and H1975/OTR cells (Figure 2D). Besides, the knockdown efficiency of siRNAs against hsa_circ_0002130 was measured by qRT-PCR. The data showed that sh-circ #1, sh-circ #2 and sh-circ #3 could significantly downregulate the expression of hsa_circ_0002130 in both HCC827/OTR and H1975/OTR cells (Figure 2E). Furthermore, sh-circ #1 possessing the best knockdown efficiency was selected for the following experiments.
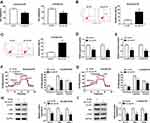
hsa_circ_0002130 Knockdown Inhibited Cell Proliferation, Glycolysis, and Enhanced Cell Apoptosis in Osimertinib-Resistant NSCLC
Next, we explored the effects of hsa_circ_0002130 on osimertinib-resistant NSCLC. MTT assay indicated that knockdown of hsa_circ_0002130 significantly inhibited cell viability in both HCC827/OTR and H1975/OTR cells (Figure 3A). hsa_circ_0002130 deletion promoted cell apoptosis in HCC827/OTR and H1975/OTR cells (Figure 3B and C). Moreover, the levels of glucose uptake and lactate production were measured. As shown in Figure 3D and E, downregulation of hsa_circ_0002130 markedly decreased the levels of glucose uptake and lactate production in both HCC827/OTR and H1975/OTR cells. Analogously, the level of ECAR was also confirmed to be downregulated in HCC827/OTR and H1975/OTR cells transfected with sh-circ #1 (Figure 3F and G). Western blot analysis suggested that downregulation of hsa_circ_0002130 significantly attenuated the expression of GLUTI, HK2 and LDHA in both osimertinib-resistant HCC827 and H1975 cells (Figure 3H and I).
Furthermore, the data in Supplement Figure 1A showed a successful overexpression transfection efficiency of oe-hsa_circ_0002130 in both HCC827/OTR and H1975/OTR cells. Overexpression of hsa_circ_0002130enhanced cell viability (Supplement Figure 1B), but had no effect on cell apoptosis in HCC827/OTR and H1975/OTR cells (Supplement Figure 1C). Besides, hsa_circ_0002130 upregulation increased the levels of glucose uptake, lactate production and ECAR (Supplement Figure 1D-G). Overexpression of hsa_circ_0002130 also elevated the protein expression of GLUT1, HK2 and LDHA (Supplement Figure 1H-I). Taken together, our results indicated that hsa_circ_0002130 knockdown could inhibit osimertinib-resistance in HCC827/OTR and H1975/OTR cells.
hsa_circ_0002130 Knockdown Suppressed Tumor Growth in vivo
To further investigate the promotion effects of hsa_circ_0002130 in osimertinib-resistant NSCLC, the nude mice injected with sh-NC/sh-circ #1 HCC827/OTR cells was treated with osimertinib. As described in Figure 4A, tumor volume was significantly inhibited by downregulating hsa_circ_0002130 in osimertinib-resistant NSCLC mice. Consistently, we also demonstrated that knockdown of hsa_circ_0002130 could significantly inhibit tumor weight (Figure 4B). Moreover, the glycolysis-related lactate production was determined. Our data showed that downregulation of hsa_circ_0002130 dramatically decreased the level of lactate production (Figure 4C). The expression of hsa_circ_0002130 was significantly repressed by hsa_circ_0002130 knockdown (Figure 4D). Then, we detected the expression of glycolysis-associated proteins including GLUT1, HK2 and LDHA. We observed that the expression of GLUT1, HK2 and LDHA was significantly suppressed by downregulating hsa_circ_0002130 (Figure 4E). All these results confirmed that hsa_circ_0002130 served as an oncogene in osimertinib-resistant NSCLC.
hsa_circ_0002130 Sponged miR-498 to Regulate GLUT1, HK2 and LDHA Expression
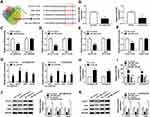
Venn diagram showed that miR-498 harbored the binding sites of hsa_circ_0002130, GLUT1, HK2 and LDHA (Figure 5A). CircInteractome predicted that hsa_circ_0002130 contained the binding sites of miR-498 and starBase predicted that miR-498 contained the binding sites of GLUT1, HK2 and LDHA (Figure 5A). The expression of miR-498 was reduced in HCC827/OTR and H1975/OTR cells compared with the HCC827 and H1975 cells (Figure 5B). Dual-luciferase reporter assay indicated that the relative luciferase activity was attenuated in HEK293T cells co-transfected with WT-hsa_circ_0002130, WT-GLUT1, WT-HK2 or WT-LDHA and miR-498 mimic compared with the corresponding control group, while the luciferase activity of MUT-hsa_circ_0002130, MUT-GLUT1, MUT-HK2 or MUT-LDHA vectors was not changed in HEK293T cells transfected with miR-498 mimic (Figure 5C–F). Meanwhile, the results demonstrated that hsa_circ_0002130, GLUT1, HK2 and LDHA were effectively enriched by miR-498 in HCC827/OTR and H1975/OTR cells (Figure 5G). The data indicated that the expression of miR-498 was significantly increased by downregulating hsa_circ_0002130 in both HCC827/OTR and H1975/OTR cells (Figure 5H). Besides, we found the successful overexpression and deletion efficiency of miR-498 mimic and inhibitor in both HCC827/OTR and H1975/OTR cells (Figure 5I). Furthermore, the expression of GLUT1, HK2 and LDHA was dramatically inhibited by upregulating miR-498 and significantly facilitated through downregulating miR-498 in osimertinib-resistant HCC827 and H1975 cells (Figure 5J and K).
miR-498 Deletion Reversed the Effects of hsa_circ_0002130 Knockdown on Osimertinib-Resistance in Osimertinib-Resistant NSCLC Cells
To further explore the underlying mechanism of hsa_circ_0002130 in osimertinib-resistant NSCLC, we performed the rescue experiments by co-transfecting sh-circ #1 and miR-498 inhibitor into HCC827/OTR and H1975/OTR cells. Cell viability of HCC827/OTR and H1975/OTR cells were significantly inhibited by miR-498 mimic, and downregulation of miR-498 reversed the inhibition effect of hsa_circ_0002130 deletion on cell viability (Figure 6A). Moreover, miR-498 upregulation significantly enhanced the number of apoptotic cells, while miR-498 deletion reversed the promotion effect of hsa_circ_0002130 downregulation on cell apoptosis (Figure 6B). Furthermore, we found that overexpression of miR-498 dramatically inhibited the levels of glucose uptake and lactate production, while the inhibitory effects of hsa_circ_0002130 downregulation on glucose uptake and lactate production were blocked by knockdown of miR-498 in both HCC827/OTR and H1975/OTR cells (Figure 6C and D). Consistently, the level of ECAR was significantly suppressed by overexpressing miR-498, while miR-498 deletion could reserve the decrease of ECAR induced by downregulating hsa_circ_0002130 (Figure 6E and F). We also demonstrated that miR-498 overexpression could inhibit the expression of GLUT1, HK2 and LDHA, while miR-498 deletion could significantly rescue the decrease of GLUT1, HK2 and LDHA expression induced by hsa_circ_0002130 downregulation (Figure 6G and H). Our results indicated that hsa_circ_0002130 was involved in the osimertinib-resistance in osimertinib-resistant NSCLC cells by combining with miR-498 to regulate the expression of GLUT1, HK2 and LDHA.
miR-498 Was Decreased, While hsa_circ_0002130, GLUT1, HK2 and LDHA Were Increased in Osimertinib-Resistant NSCLC Patients
Our data showed that the expression of miR-498 was downregulated, while hsa_circ_0002130, GLUT1, HK2 and LDHA were upregulated in osimertinib-resistant NSCLC patients (Figure 7A–E). Moreover, hsa_circ_0002130 expression was negatively related to miR-498 in NSCLC patients (Figure 7F). Furthermore, hsa_circ_0002130 expression was positively associated with the expression of GLUT1, HK2 or LDHA (Figure 7G–I).
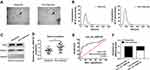
hsa_circ_0002130 Was Secreted by Exosomes from the Serum of NSCLC Patients
Finally, in the present study, the abundant sera were collected from 28 non-response NSCLC patients and 32 response NSCLC patients. The isolated exosomes using sequential centrifugation were verified using electron microscopy (Figure 8A). To confirm the purification of exosomes, the vesicles were determined by Nanosight NTA. The results of NTA analysis indicated that the diameter of the vesicles ranged from 30 to 140 nm that is in line with the size of exosomes (Figure 8B). Western blot confirmed the presence of two well-known exosomal markers, CD81 and TSG101 (Figure 8C). QRT-PCR analysis showed that hsa_circ_0002130 was enriched in serum exosomes derived from osimertinib-resistant NSCLC patients compared with that in osimertinib-sensitive NSCLC patients (Figure 8D). Moreover, we demonstrated that hsa_circ_0002130 had the diagnostic value of AUC=0.792 (P<0.001) (Figure 8E). As shown in Figure 8F, the proportion of non-response NSCLC patients in NSCLC patients with high hsa_circ_0002130 expression was higher than that in NSCLC patients with low hsa_circ_0002130 expression. All data demonstrated that serum exosomal hsa_circ_0002130 was upregulated in osimertinib-resistant NSCLC patients.
Discussion
Cancer drug-resistance is still a difficult problem in human cancer clinical treatment. Osimertinib is an effective EGFR-tyrosine kinase inhibitor for the advanced NSCLC patients, yet it is also inevitable to acquire resistance to osimertinib.27 Previous studies suggested that cancer cells exerted facilitated effect on glycolysis for ATP generation, which promoted cancer development and progression.28 Several studies indicated that glycolysis potently inhibited the apoptotic rate of multidrug-resistant cells, suggesting that a novel strategy of targeting glycolysis might be useful to overcome multidrug resistance.29 In our study, we found that the levels of glucose uptake, lactate production and ECAR were significantly increased in osimertinib-resistant NSCLC cells. Moreover, the glycolysis-related proteins, including GLUT1, HK2 and LDHA, were dramatically upregulated in osimertinib-resistant NSCLC cells. Our data indicated that glycolysis exhibited a promotion role in osimertinib-resistant NSCLC.
In the recent few years, noncoding RNAs, such as circRNAs and miRNAs, have been confirmed to be involved and played vital roles in verities of human pathophysiologic processes including tumorigenesis.30 Despite the growing evidence is usable to verify the underlying mechanisms of circRNAs in the progression of malignant tumors, the knowledge is still limited. CircRNAs, such as circ_100876, circ_CCDC66 and circ_ITCH, were confirmed to be associated with development, lymph node metastasis, pathological grade, as well as drug resistance in lung cancer.31–33 In this paper, we discovered that hsa_circ_0002130 was highly expressed in osimertinib-resistant NSCLC cells and knockdown of hsa_circ_0002130 could significantly exert inhibitory effects on cell growth and glycolysis, but exhibit an enhanced effect on cell apoptosis of osimertinib-resistant NSCLC cells. Moreover, hsa_circ_0002130 deletion also exerted inhibition effects on tumor growth in vivo. hsa_circ_0002130 might be a novel treatment target for osimertinib-resistant NSCLC. Consistently, it has been discovered that hsa_circ_0002130 was also upregulated in ovarian endometriosis and pancreatic ductal adenocarcinoma.34,35
Accumulating evidence demonstrated that circRNAs, a novel group of noncoding RNAs, served as miRNA sponges to regulate the downstream gene expression.36 For example, circ-ZFR could sponge miR-30a/miR-107 to modulate PTEN expression in gastric cancer, while circLARP4 acted as an inhibitor in gastric cancer progression via targeting miR-424-5p and regulating the expression of LATS1.37,38 Liu and his colleagues indicated that hsa_circRNA_103809 facilitated the development of lung cancer through sequestering miR-4302 to enhance the expression of MYC, suggesting that hsa_circRNA_103809 might be a prognosis biomarker for lung cancer patients.39 Furthermore, circ-PRMT5 boosted cell growth of NSCLC cells via increasing the expression of EXH2 through sponging miR-377/382/498.40 More importantly, accumulating evidence revealed that circRNAs could regulate gene expression via targeting miRNAs in drug-resistant cancers.41 Consistently, we found that hsa_circ_0002130 could target miR-498 to regulate the expression of glycolysis-related proteins, including GLUT1, HK2 and LDHA in osimertinib-resistant NSCLC cells. In our research, miR-498 overexpression significantly suppressed cell growth and glycolysis, and enhanced the apoptotic ability of osimertinib-resistant NSCLC cells. Moreover, our data revealed that the suppressive effects of hsa_circ_0002130 deletion were reversed by downregulating miR-498 in osimertinib-resistant NSCLC cells.
Exosomes are membrane vesicles released by various cells into the extracellular microenvironment.42 Accumulating evidence suggested that exosomes might be crucial mediators in types of human cancers, which can participate in the intercellular communication through transferring circRNAs, miRNAs, mRNAs and proteins.43 A previous research indicated that exosomal circRNAs acted as molecular biomarkers for the monitoring, prognosis and diagnosis of diseases, including human cancer.44 For instance, exosomal circ-PDE8A facilitated tumor growth and invasive in pancreatic cancer.45 Notably, we discovered that the enhanced expression of hsa_circ_0002130 could be determined in the serum exosomes of osimertinib-resistant NSCLC patients.
In summary, hsa_circ_0002130 expression was significantly enhanced in osimertinib-resistant NSCLC cells and serum exosomes from osimertinib-resistant NSCLC patients, which exerted promotion effects on osimertinib-resistance. Furthermore, downregulating hsa_circ_0002130 dramatically suppressed the progression of osimertinib-resistant NSCLC both in vitro and in vivo. hsa_circ_0002130 affected cell growth, apoptosis and glycolysis by sponging miR-498 to regulate GLUT1, HK2 and LDHA in osimertinib-resistant NSCLC cells.
Disclosure
The authors declare that they have no financial conflicts of interest.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
3. Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6(2):244–285. doi:10.1097/JTO.0b013e318206a221
4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
5. Wang B, Zuo Z, Li F, Yang K, Du M, Gao Y. Gefitinib versus docetaxel in treated non-small-cell lung cancer: a meta-analysis. Open Med. 2017;12:86–91. doi:10.1515/med-2017-0013
6. Bollinger MK, Agnew AS, Mascara GP. Osimertinib: a third-generation tyrosine kinase inhibitor for treatment of epidermal growth factor receptor-mutated non-small cell lung cancer with the acquired Thr790Met mutation. J Oncol Pharm Pract. 2018;24(5):379–388. doi:10.1177/1078155217712401
7. Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. doi:10.1158/1078-0432.CCR-12-2246
8. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi:10.1038/nrm.2015.32
9. Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75(6):1071–1098. doi:10.1007/s00018-017-2688-5
10. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
11. Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6(5):563–579. doi:10.1002/wrna.1294
12. Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5(2):472–480.
13. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
14. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
15. Han W, Wang L, Zhang L, Wang Y, Li Y. Circular RNA circ-RAD23B promotes cell growth and invasion by miR-593-3p/CCND2 and miR-653-5p/TIAM1 pathways in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;510(3):462–466. doi:10.1016/j.bbrc.2019.01.131
16. Chen T, Luo J, Gu Y, Huang J, Luo Q, Yang Y. Comprehensive analysis of circular RNA profiling in AZD9291-resistant non-small cell lung cancer cell lines. Thorac Cancer. 2019;10(4):930–941. doi:10.1111/1759-7714.13032
17. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi:10.1126/science.1160809
18. Liu Y, Cao Y, Zhang W, et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol Cancer Ther. 2012;11(8):1672–1682. doi:10.1158/1535-7163.MCT-12-0131
19. Agnihotri S, Zadeh G. Metabolic reprogramming in glioblastoma: the influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol. 2016;18(2):160–172. doi:10.1093/neuonc/nov125
20. Marin-Valencia I, Yang C, Mashimo T, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15(6):827–837. doi:10.1016/j.cmet.2012.05.001
21. Ooi AT, Gomperts BN. Molecular Pathways: targeting Cellular Energy Metabolism in Cancer via Inhibition of SLC2A1 and LDHA. Clin Cancer Res. 2015;21(11):2440–2444. doi:10.1158/1078-0432.CCR-14-1209
22. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8(1):83. doi:10.1186/s13045-015-0181-x
23. Li C, Xu X. Biological functions and clinical applications of exosomal non-coding RNAs in hepatocellular carcinoma. Cell Mol Life Sci. 2019;76(21):4203–4219. doi:10.1007/s00018-019-03215-0
24. Santangelo L, Giurato G, Cicchini C, et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17(3):799–808. doi:10.1016/j.celrep.2016.09.031
25. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi:10.1038/cr.2015.82
26. Muller L, Mitsuhashi M, Simms P, Gooding WE, Whiteside TL. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci Rep. 2016;6(1):20254. doi:10.1038/srep20254
27. Cross DA, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4(9):1046–1061. doi:10.1158/2159-8290.CD-14-0337
28. Menendez JA, Alarcon T. Metabostemness: a new cancer hallmark. Front Oncol. 2014;4:262. doi:10.3389/fonc.2014.00262
29. Xu RH, Pelicano H, Zhou Y, et al. Inhibition of glycolysis in cancer cells: a novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res. 2005;65(2):613–621.
30. Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40(4):902–906. doi:10.1042/BST20120020
31. Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH Suppresses lung cancer proliferation via inhibiting the wnt/beta-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi:10.1155/2016/1579490
32. Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456. doi:10.1016/j.prp.2017.02.011
33. Ide M, Kato T, Ogata K, Mochiki E, Kuwano H, Oyama T. Keratin 17 expression correlates with tumor progression and poor prognosis in gastric adenocarcinoma. Ann Surg Oncol. 2012;19(11):3506–3514. doi:10.1245/s10434-012-2437-9
34. Li Q, Geng S, Yuan H, et al. Circular RNA expression profiles in extracellular vesicles from the plasma of patients with pancreatic ductal adenocarcinoma. FEBS Open Bio. 2019;9(12):2052–2062. doi:10.1002/2211-5463.12741
35. Wang D, Luo Y, Wang G, Yang Q. Circular RNA expression profiles and bioinformatics analysis in ovarian endometriosis. Mol Genet Genomic Med. 2019;7(7):e00756. doi:10.1002/mgg3.756
36. Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18(5):780–788. doi:10.1093/bib/bbw053
37. Liu T, Liu S, Xu Y, et al. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res Treat. 2018;50(4):1396–1417. doi:10.4143/crt.2017.537
38. Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi:10.1186/s12943-017-0719-3
39. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi:10.1016/j.bbrc.2018.04.172
40. Wang Y, Li Y, He H, Wang F. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene. 2019;720:144099. doi:10.1016/j.gene.2019.144099
41. Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18(1):71. doi:10.1186/s12943-019-0969-3
42. Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101(3):942–948. doi:10.1083/jcb.101.3.942
43. Wang J, Hendrix A, Hernot S, et al. Bone marrow stromal cell-derived exosomes as communicators in drug resistance in multiple myeloma cells. Blood. 2014;124(4):555–566. doi:10.1182/blood-2014-03-562439
44. Fanale D, Taverna S, Russo A, Bazan V. Circular RNA in Exosomes. Adv Exp Med Biol. 2018;1087:109–117. doi:10.1007/978-981-13-1426-1_9
45. Li Z, Yanfang W, Li J, et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi:10.1016/j.canlet.2018.04.035
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.