Back to Journals » International Journal of Nanomedicine » Volume 13
A novel role in skeletal segment regeneration of extracellular vesicles released from periodontal-ligament stem cells
Authors Diomede F , D'Aurora M, Gugliandolo A, Merciaro I, Ettorre V, Bramanti A , Piattelli A, Gatta V, Mazzon E, Fontana A , Trubiani O
Received 17 January 2018
Accepted for publication 14 March 2018
Published 29 June 2018 Volume 2018:13 Pages 3805—3825
DOI https://doi.org/10.2147/IJN.S162836
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Francesca Diomede,1 Marco D’Aurora,2 Agnese Gugliandolo,3 Ilaria Merciaro,1 Valeria Ettorre,4 Alessia Bramanti,3,5 Adriano Piattelli,1 Valentina Gatta,2 Emanuela Mazzon,3 Antonella Fontana,4 Oriana Trubiani1
1Department of Medical, Oral, and Biotechnological Sciences, University “G. d’Annunzio”, Chieti, Italy; 2Department of Psychological, Health, and Territorial Sciences, University “G. d’Annunzio”, Chieti, Italy; 3Department of Experimental Neurology, IRCCS Centro Neurolesi “Bonino Pulejo”, Messina, Italy; 4Department of Pharmacy, University “G. d’Annunzio”, Chieti, Italy; 5Eduardo Caianiello Institute of Applied Science and Intelligent Systems (ISASI), National Research Council, Messina, Italy
Purpose: The combination of oral derived stem cells and 3-D scaffolds is considered advantageous in bone repair. In particular, collagen membranes possess ideal biological properties and can support infiltration and proliferation of osteoblasts, promoting bone regeneration. Our study aimed to develop a new biocompatible osteogenic construct composed of a commercially available collagen membrane (Evolution [Evo]), human periodontal-ligament stem cells (hPDLSCs) enriched with extracellular vesicles (EVs), or polyethylenimine (PEI)-engineered EVs (PEI-EVs).
Methods: Osteogenic ability and expression of osteogenic genes were evaluated in vitro in hPDLSCs cultured with or without Evo, with Evo and EVs, or PEI-EVs. In addition, the bone-regeneration capacity of Evo, Evo enriched with hPDLSCs, Evo enriched with hPDLSCs and EVs/PEI-EVs was investigated in rats subjected to calvarial defects.
Results: Our results showed that Evo enriched with EVs and PEI-EVs showed high biocompatibility and osteogenic properties in vitro and in vivo. In addition, quantitative reverse-transcription polymerase chain reaction demonstrated the upregulation of osteogenic genes, such as TGFB1, MMP8, TUFT1, TFIP11, BMP2, and BMP4, in the presence of PEI-EVs. Upregulation of BMP2/4 was confirmed for Evo enriched with PEI-EVs and hPDLSCs both in vitro by Western blot and in vivo by immunofluorescence.
Conclusion: Our results indicated that Evo enriched with hPDLSCs and PEI-EVs is able to promote a bone-regeneration process for the treatment of calvarium and ossification defects caused by accidental or surgery trauma. In particular, PEI-EVs had a significant role in activation of the osteogenic process.
Keywords: human periodontal-ligament stem cells, living construct, extracellular vesicles, bone regeneration, collagen membrane
Introduction
Bone is a specialized, highly dynamic connective tissue, with osteoblasts, osteoclasts, and blood vessels involved in the maintenance of its homeostasis.1 In bone regeneration, the use of biomaterials is aimed at providing a provisional three-dimensional scaffold and mechanical support to the cells.2 An ideal biomaterial should be osteogenic, ie, with the capacity to induce bone-tissue formation, osteoinductive for the possibility to recruit mesenchymal stem cells (MSCs) derived from the host, with bioactive effects on ossification, and osteoconductive, providing a three-dimensional scaffold for the ingrowth of vessels and osteoprogenitor cells. Moreover, it should be biocompatible, resorbable, affordable, and easy to use. In particular, collagen membranes show great biocompatibility and are able to support bone regeneration, and for this reason they are widely employed for bone-tissue regeneration3,4 and to augment deficient alveolar-ridge volume.5
The combination of biomaterials and SCs represents a common strategy for bone-tissue-engineering applications. In particular, oral tissue represents an easily accessible source of SCs that can be used in regenerative medicine.6 In this study, we focused on human periodontal-ligament stem cells (hPDLSCs), which show (similarly to other MSCs) in vitro proliferative ability and multilineage-differentiation capacity.7 Indeed, hPDLSCs have been reported to differentiate into osteogenic, adipogenic, chondrogenic, and neurogenic cell lineages in vitro.8,9 Moreover, hPDLSCs have shown the ability to promote the bone-regenerative process in association with different scaffolds.10,11 In addition, hPDLSCs exert immunomodulatory12 and therapeutic effects, which may be mediated at least in part through the release of extracellular vesicles (EVs) that act as paracrine signalers.13 Exosomes are small well-defined vesicles, ie, phospholipidic membrane-enclosed entities containing cytokines, proteins, lipids, and nucleic acids, such as mRNA and microRNAs. Exosomes derived from MSCs show great potential in different fields of regenerative medicine, ranging from neural to skeletal regeneration.14,15 A study in vitro demonstrated that EVs derived from bone-marrow MSCs (BMSCs) were able to enter the osteoblast and positively regulate osteoblastic differentiation.16 In line with this result and also in vivo studies, EVs derived from different types of MSCs, including BMSCs and embryonic MSCs, have been reported to facilitate the processes of fracture healing, osteochondral regeneration, and calvarial bone-defect repair.16–18
With the aim of improving scaffold performance, we enriched it with SCs and EVs or engineered EVs to evaluate the bone-regenerative properties of these biocomplexes. For the engineering of EVs, polyethylenimine (PEI) was used. PEI is a synthetic polymer with high cationic charge density, due to the presence of protonatable amino groups.19 PEI is widely used to deliver DNA molecules20 and small oligonucleotides,19,21–23 both in vitro and in vivo. In particular, in vitro data demonstrate low toxicity and high biological activity for PEI.24 This effect is likely due to PEI capacity to form nonconvalent complexes with the DNA, which can be efficiently taken up by cells through endocytosis.24 Another important capability of PEI is its ability to induce the intracellular release of PEI–nucleic acid complexes from endosomes, thanks to the induction of osmotic swelling (ie, proton-sponge effect) that causes the burst of endosomes without the need for an additional endosomolytic agent.25 Therefore the engineering of EVs (PEI-EVs), obtained by noncovalently coating vesicles with PEI, should promote the release of EV content into cells.
The aim of this study was to investigate the properties of collagen membrane-engineered scaffolds enriched with hPDLSCs and EVs or with hPDLSCs and PEI-EVs in bone regeneration through in vitro and in vivo evaluations. We evaluated the ability of the different scaffolds to induce osteogenic differentiation and modulate the expression of genes involved in bone processes in vitro and induce bone regeneration in vivo in rats subjected to calvarial defects. Especially, we were interested in comparing the therapeutic potential of EVs and PEI-EVs, in order to assess their contribution to bone regeneration. The final purpose of this study was the development of a new biocompatible and osteogenic construct useful for bone repair, especially of calvarial defects. To our knowledge, this is the first study to evaluate the bone-regenerative capacity of these constructs formed by collagen-membrane scaffolds enriched with hPDLSCs and EVs or PEI-EVs.
Materials and methods
Scaffold material
The Evolution (Evo; Tecnoss Dental, Giaveno, Italy) membrane is a high-consistency dense collagen fiber derived from equine mesenchymal tissue. It shows up as a sterile dried membrane with smooth and microrough sides (Figure S1A and B). The major characteristics of this material are its maximum adaptability to hard and soft tissue, easy and secure suturability of nearby tissue, ample stability, and sufficient protection of underlying grafts. Indicated mainly for surgical procedures, Evo can also be used as a drug carrier. Sterile scissors were used to obtain the desired piece size. Evo membrane pieces were washed with sterile PBS (LiStarFish, Milan, Italy) to rehydrate the material before use.
hPDLSCs ex vivo patient ethics statement
Written approval for the hPDLSCs collection performed in this study was obtained from the medical ethics committee at the Medical School, University “G. d’Annunzio”, Chieti, Italy (266/17.04.14). Written informed consent for clinical research and for processing of personal data was obtained from all subjects before sample collection. The Department of Medical, Oral, and Biotechnological Sciences, University “G. d’Annunzio”, and the Laboratory of Stem Cells and Regenerative Medicine, University “G. d’Annunzio”, are certified according to the quality standard ISO 9001:2008 RINA (certificate 32031/15/S).
In vitro analysis
Cell culture
Five persons, either patients for orthodontic purposes or healthy volunteers, were selected for tooth removal. Cells were collected as previously described by Diomede et al.26 Subsequently, cells were cultivated using the nonchemically defined MSCGM-CD BulletKit medium (Lonza, Basel, Switzerland), which was changed twice a week, to allow the growth of human MSCs and minimize exposure to nonhuman substances.27 hPDLSC control cells are shown in Figure S1D.
hPDLSC cytofluorometric characterization
Flow-cytometry analysis was performed on hPDLSCs at second passage as previously described by Rajan et al.28 Data were analyzed using FlowJo (Ashland, OR, USA) software.
Morphological analysis of hPDLSCs
To evaluate plastic adherent ability, hPDLSCs at P2 were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 2 hours, subsequently stained with toluidine blue and observed by light microscopy (DMIL; Leica Microsystems, Wetzlar, Germany).
Mesengenic differentiation ability of hPDLSCs
To assay the ability of hPDLSCs to differentiate into mesengenic lineages, cells were cultured in osteogenic and adipogenic induction/maintenance medium (Lonza). Osteogenic and adipogenic capability was assayed using Alizarin red S and oil red staining as previously described by Ballerini et al29 and observed by light microscopy.
hPDLSC extracellular vesicle isolation
After 48 hours of incubation, the conditioned medium (CM; 10 mL), was collected from hPDLSCs at second passage. The CM was centrifuged at 3,000 g for 15 minutes to eliminate suspension cells and debris. For EV extraction, ExoQuick TC commercial agglutinant (System Biosciences, Palo Alto, CA, USA) was used. Briefly, 2 mL ExoQuick TC was added to 10 mL CM recovered from hPDLSCs. The mix was incubated overnight at 4°C without rotation, one centrifugation step was performed at 1,500 g for 30 minutes to sediment the EVs, and pellets were resuspended in 200 μL PBS. Split into two aliquots, EVs were precipitated, and quantification of whole-homogenate proteins was used as confirmation of the presence of EV release in hPDLSCs.13
Engineered EV preparation
EVs were engineered by noncovalently coating EVs with PEI.30,31 EV pellets (100 μL) were resuspended in 2 mL PBS. Branched PEI solution (2 mL, molecular weight 25,000 Da; Sigma-Aldrich, St Louis, MO, USA) in 0.3 M NaCl was added to the EVs suspension in PBS and the mixture incubated for 20 minutes at room temperature. We tested different concentrations of PEI (0.025, 0.5, 0.1, 0.05, and 5 mg/mL), to find the best compromise between activity and toxicity. Then, the suspension was centrifuged at 4,000 rpm for 15 minutes and the supernatant removed to get rid of the excess PEI. The precipitate was resuspended in 2 mL PBS. The PEI-EV suspension was characterized using dynamic light-scattering experiments and ζ-potential measurements.
To evaluate interaction between the PEI-EVs and hPDLSCs, wheat-germ agglutinin (WGA) Alexa Fluor 488-stained PEI-EVs were analyzed by confocal laser-scanning microscopy (CLSM) after 24 hours of incubation. To evaluate cell viability, hPDLSCs were cultured in the presence of EVs complexed with different concentrations of PEI (0.025, 0.05, 0.1, 0.5, and 5 mg/mL) for 24 hours. Cells were seeded at a density of 103 cells/well in triplicate using a 96-well flat-bottom plate and maintained for 24, 48, and 72 hours and 1 week. Trypan blue solution (0.1%) was added to the cell suspension for 3–5 minutes. The number of dead cells and living cells in total cells were then counted.32
Atomic force-microscopy measurements
To evaluate EVs and PEI-EVs, atomic force-microscopy measurements were performed using a Multimode 8 microscope with NanoScope V controller (Bruker, Billerica, MA, USA). Silicon cantilever and a RTESPA-300 tip (spring constant 40 N/m and resonance frequency 300 Hz) were used in tapping mode. Specimens were prepared by dropping a solution of EVs and PEI-EVs onto an SiO2 wafer, followed by air-drying at 37°C for 1 hour. Since it was necessary to remove the solvent to perform this analysis, the PEI-EV solution was diluted to reduce PEI-EV tendency to aggregate on the SiO2 wafer.
Evo, EV, and hPDLSC interaction
To evaluate the capacity of EVs and PEI-EVs to adhere onto Evo membrane, EVs and PEI-EVs were stained with Alexa Fluor 488 and subsequently seeded on the membrane for 24 hours and maintained at 37°C. After 24 hours of incubation, all samples were analyzed with CLSM (LSM800; Carl Zeiss, Jena, Germany).33
Spontaneous osteogenic differentiation
hPDLSCs at second passage were seeded at 8×103 cells/cm2 and cultured in MSCGM-CD medium in the presence of Evo, Evo + EVs, and Evo + PEI-EVs. Briefly, sample groups were divided: hPDLSCs, Evo + hPDLSCs, Evo + EVs + hPDLSCs, and Evo + PEI-EVs + hPDLSCs. After 6 weeks, visualization of calcium deposition and extracellular matrix mineralization was obtained by Alizarin red S staining assay, in accordance with Trubiani et al.34
RNA extraction and TaqMan quantitative real-time polymerase chain reaction
Total RNA was extracted using a RNeasy minikit (Qiagen, Venlo, the Netherlands). A High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used for reverse transcription of 6 μg RNA from each sample. Quantitative real-time polymerase chain reaction (PCR) was performed on a 96-well TaqMan Array human-osteogenesis kit following the manufacturer’s instructions and run on an ABI 7900HT sequencing-detection system (Thermo Fisher Scientific). The array contains 92 assays related to osteogenic differentiation, including transcription and growth factors, and genes involved in the physiological processes of bone and tooth formation, mineralization, and maintenance. Amplification conditions were 10 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. Three independent experiments were run for each condition, for a total of 12 plates. Real-time data were analyzed with DataAssist software (Thermo Fisher Scientific). ΔΔCt relative global expression analysis was used and GAPDH, 18S, GUSB, and HPRT1 were chosen as selected endogenous controls. Only genes showing a maximum Ct value of 35 and no outliers or replicates were included in the analysis. A gene was considered differentially expressed if showing a fold change >1.2 or <0.7 and a P-value <0.05. P-values were adjusted using Benjamini–Hochberg false-discovery rate. Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood, CA, USA) was employed to infer biological functions and networks for the resulting gene data sets. IPA predicts functional networks based on known gene-function interactions and ranks them by significance.
Western blot analysis
Protein extraction and Western blot were performed as previously reported.35,36 BMP2/4 (1:500; Santa Cruz Biotechnology, Dallas, TX, USA) was used as primary antibody. β-Actin (1:750; Santa Cruz Biotechnology) was used to assess uniform protein loading. Bands were analyzed by enhanced luminescence using Alliance 2.7 (Uvitec, Cambridge, UK).
In vivo analysis
Animals
Male Wistar rats, weighing 300–350 g, were used for this experiment. Animals were acquired from Harlan Laboratories (Indianapolis, IN, USA), housed in individually ventilated cages, and maintained under 12-hour light/dark cycles at 21°C±1°C and 50%–55% humidity with food and water ad libitum.
Ethics statement for animal use
All animal care, and use, complied with the European Union (EU) guidelines for animal welfare. The study was authorized by the Ministry of Health, Italy (“General direction of animal health and veterinary drugs”, authorization 768/2016-PR 28/07/2016). Experiments were planned to minimize the total number of rats needed for the study.
Scaffold implants
To implant scaffolds, rats were first anesthetized with a combination of tiletamine and xylazine (10 mL/kg intraperitoneally). Afterward, the implant site was prepared with povidone–iodine (Betadine®). After trichotomy, a median sagittal incision of about 1 cm in the frontoparietal region was performed, calvaria exposed, and a circular section of the bone-receiving site (diameter 4 mm, height 0.25 mm) was injured with a dedicated rotary instrument at a controlled speed (trephine milling machine; Alpha Bio Tec, Petach Tikva, Israel) under constant irrigation with a physiological solution. Due to their texture and flexibility, Evo, Evo + hPDLSCs, Evo + EVs, Evo + EVs + hPDLSCs, Evo + PEI-EVs, and Evo + PEI-EVs + hPDLSCs were inserted easily into contact with bone tissue to cover the damaged area. The skin flap was then sutured with small absorbable sutures of reduced diameter (Caprosyn 6-0), using interrupted points. Standard feeding and hydration were maintained constantly throughout the postoperative phase.
Experimental design
Rats were randomly distributed into the following groups (n=24 total animals):
- Evo (n=4): rats subjected to scraping of cortical calvarium bone tissue and implant of Evo.
- Evo + hPDLSCs (n=4): rats subjected to scraping of cortical calvarium bone tissue and implant of Evo enriched with hPDLSCs.
- Evo + EVs (n=4): rats subjected to scraping of cortical calvarium bone tissue and implant of Evo enriched with EVs.
- Evo + EVs + hPDLSCs (n=4): rats subjected to scraping of cortical calvarium bone tissue and implant of Evo enriched with EVs + hPDLSCs.
- Evo + PEI-EVs (n=4): rats subjected to scraping of cortical calvarium bone tissue and implant of Evo enriched with PEI-EVs.
- Evo + PEI-EVs + hPDLSCs (n=4): rats subjected to scraping of cortical calvarium bone tissue and implant of Evo enriched with PEI-EVs + hPDLSCs.
After 6 weeks, the animals were euthanized by intravenous administration of Tanax (5 mL/kg body weight) and their calvaria processed for morphological analysis.
Preparation of hPDLSCs, EVs, and PEI-EVs for in vivo experiments
For each experimental condition, 2×106 hPDLSCs were labeled for 30 minutes with the red fluorescent lipophilic dye PKH26 (Sigma-Aldrich) according to the manufacturer’s instructions. PKH26 is stably incorporated in lipid regions of the cell membrane, allowing in vivo cell tracking and monitoring studies. PKH26-labeled hPDLSCs were seeded on Evo and then incubated for 24 hours at 37°C in 5% CO2 and air before implantation into rat calvaria. EVs and PEI-EVs were also stained with PKH76.
Histological evaluation
The specimens were fixed for 72 hours in 10% formalin solution, dehydrated in ascending graded alcohol, and embedded in London Resin white resin (Sigma-Aldrich). After polymerization, undecalcified oriented cut sections of 100 μm were prepared and ground down to about 80 μm using the TT system (TMA 2, Grottammare, Italy). Sections were analyzed with an LSM 510 Meta (Zeiss), and after staining with a solution of acid fuchsine and methylene blue, observed with light microscopy. Investigation was carried out by means of bright-field light microscopy connected to a high-resolution digital camera (DFC425B; Leica Microsystems). Three-dimensional reconstruction was obtained by means of ZEN2 software (Zeiss). Statistical analysis was performed using SPSS version 21.0.
Immunohistochemistry and CLSM analysis
Semithin undecalcified sections embedded in London Resin white resin of all samples were processed for immunofluorescence labeling. Samples were blocked with 3% bovine serum albumin (BSA) in PBS-Tween for 1 hour. Primary monoclonal antibody antihuman BMP2/4 (1:100, mouse) was used, followed by Alexa Fluor 488 green fluorescence-conjugated goat antimouse. Sections were analyzed with CLSM (LSM800). The investigation was carried out with bright-field light microscopy connected to a high-resolution digital camera (DFC425B).
Data and statistical analysis
SPSS version 21.0 was used for data analysis. Parametrical methods were used after verifying the existence of the required assumptions. In particular, the normality of the distribution and equality of variance were assessed by the Shapiro–Wilk and Levene tests, respectively. Data are expressed as mean ± standard deviation. Differences among groups were determined using Student’s t-test. Differences were considered significant at P<0.05.
Results
hPDLSC characterization
Cytofluorometric evaluation of hPDLSCs showed high positivity for CD13, CD29, CD44, CD73, CD90, CD105, CD146, and CD166, while negative expression of CD14, CD34, CD45, CD117, CD133, CD144, and CD271 was present (Figure S1C). Plastic adherent hPDLSCs at second passage showed fibroblastic morphology with elliptical nuclei (Figure S1D). Osteogenic differentiation of hPDLSCs after 21 days under induction conditions was evidenced with Alizarin red staining (Figure S1E). Oil red O solution was used to evaluate adipogenic commitment. Lipid droplets at cytoplasmic level were stained in red (Figure S1F).
PEI-EV characterization
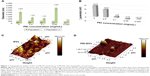
Dynamic light-scattering analysis evidenced the presence of two main dimensional populations of vesicles, namely population 1 and population 2, with an average diameter of 90±20 nm and 1,200±400 nm, respectively. Despite in vitro and in vivo measurements being performed using a mixture of both populations, we preferred to characterize them separately, to evaluate differences, if any, occurring during the engineering process. In agreement with the engineering, both populations increased in size after the addition of PEI. In particular, PEI-EVs obtained with higher concentrations of PEI were larger than those obtained with lower concentrations of PEI. The 0.05 mg/mL PEI sample did not follow this trend, with final sizes of 250±50 nm and 3,600±500 nm for population 1 and population 2, respectively (Figure 1A). Further confirmation of coating of EVs with PEI was provided by analysis of ζ-potential values. ζ-Potentials increased on passing from EVs to PEI-EVs, and the increase was related to the PEI concentration used in the engineering process (Figure 1B). In particular, for the 0.05 mg/mL sample used for subsequent in vivo investigations, a ζ-potential of −1.2±0.9 mV was obtained. This value, relatively close to zero, could explain the exceptionally high dimensions seen for PEI-EVs obtained with 0.05 mg/mL PEI. As a matter of fact, the decreased electrostatic repulsions, due to the decrease of ζ-potential, could favor PEI-EV aggregation.
Dynamic light scattering was used for evaluating the stability of EVs over time. After 1 day, the size of EVs in PBS did not change significantly, remaining within experimental error. PEI-EVs tended to aggregate in PBS, almost doubling in size. Nevertheless, no evidence of aggregation was seen once PEI-EVs had been deposited onto Evo membranes. EVs and PEI-EVs were analyzed using atomic force microscopy in tapping mode. A large number of globular EVs of different dimensions characterized by the presence of a central depression were monitored (Figure 1C), using methods reported previous research.37 Little debris or aggregated vesicles were also visualized. Interestingly, the surface of EVs appeared relatively smooth. On the other hand, PEI-EVs (Figure 1D) showed objects of homogeneous dimensions without the central depression and with a less smooth surface with respect to pure EVs, likely due to the adsorption of PEI. EVs and PEI-EVs had different concentrations of EVs. To avoid aggregation of PEI-EVs that were more prone to self-aggregate due to their low ζ-potential on SiO2 wafer, we diluted the original solution with respect to the nonengineered EVs.
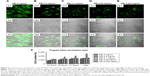
hPDLSC and PEI-EV interaction
PEI-EVs were stained with WGA Alexa Fluor 488 and incubated with hPDLSCs for 24 hours at different PEI concentrations. CLSM images were captured and cell viability estimated by means of trypan blue exclusion test (Figure 2). CLSM images showed the presence of PEI-EVs at the cytoplasmic level. In particular, EVs complexed with 0.05 mg/mL PEI concentration showed better results in terms of cell viability compared to the other PEI concentrations (P<0.01). PEI 0.05 mg/mL was chosen for subsequent experiments, as suggested by trypan blue results.
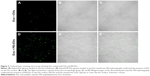
Evo, EV, and hPDLSC interaction
To evaluate the possibility of EVs and PEI-EVs of adhering to the Evo surface, CLSM images were obtained using the Zeiss LSM800. Microphotography confirmed the presence of EVs conjugate with green fluorescence on the membrane surface before grafting on rat-calvarium implant sites. PEI-EVs and EVs showed the same capacity to cover the collagen surface (Figure 3).
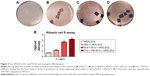
Osteogenic differentiation
Macrophotographs were performed to evaluate the alizarin red S staining at low magnification (Figure 4A–D). After 6 weeks of culture, the best results in terms of production of calcium deposition were evaluated in Evo + PEI-EVs + hPDLSCs. Data were quantified using spectrometry analysis (Figure 4E).
Genes and network analysis
In order to test Evo capacity to induce the expression of human osteogenic genes in hPDLSCs, a gene-expression profile of hPDLSCs in the presence of Evo versus hPDLSCs was analyzed (Figure 5). Quantitative reverse-transcription PCR (qRT-PCR) of 92 osteogenesis-related transcripts was performed to evidence how the gene-expression profile was modulated by Evo when compared to cells differentiated without biomaterial. Evo + hPDLSCs induced the modulation of 40 genes (Figure 5), with only MMP8 being upregulated. Cells cultured in the presence of Evo + EVs showed the modulation of 36 genes (Figure 6). The enrichment of Evo with EVs compared with hPDLSCs determined an overall downward expression modulation, whereas only two genes were up-regulated: TGFB1 and TGFB2. Cells cultured with Evo + PEI-EVs showed modulation of 40 transcripts (Figure 7). IPA functional analysis of the three gene data sets for each condition evidenced the involvement of peculiar biological functions related to connective and skeletal-tissue development and functions (Figures S2A, S3A, and S4A). IPA network analysis generated a top network for each data set, with scores of 48, 50, and 47 (Figures S2B, S3B, and S4B, respectively). Evo enriched with hPDLSCs induced overall downregulation when compared to hPDLSCs. Compared to Evo enriched with hPDLSCs, the presence of Evo + EVs determined the downregulation of SMAD and the upregulation of TGFB1, which are crucial factors driving pathway modulation, collagens and SMADs being the two cores of the network. The presence of Evo + PEI-EVs determined the overexpression of more genes, TGFB1, BMP2, and BMP4 the drivers of the expression modulation found. Moreover, all the conditions analyzed have RUNX2 and SOX9 strongly reduced compared to the control.
In vivo characterization
Macroscopic evaluation at 6 weeks postoperatively showed better regeneration capacity of Evo + PEI-EVs + hPDLSCs (Figure 8). At 6 weeks after grafting in the Evo-transplantation group, an extracellular matrix placed between native bone and membrane was found at the grafting sites (Figure 9A). In Evo + hPDLSCs, better integration was visible, especially at high magnification (40×). In particular, Evo + hPDLSCs showed good integration in the contact area with vascularized extracellular matrix around the native bone, with no inflammatory reaction (Figure 9B).
Harvested samples grafted with Evo + EVs + hPDLSCs were compared with samples grafted with Evo + EVs (Figure 9C). In both samples, the membrane was well integrated with host tissue, and Evo + EVs + hPDLSCs showed an osteoid formation with osteoblast-like structure on the native bone side, new bone appearing with an irregular arrangement (Figure 9D). With Evo + PEI-EVs + hPDLSCs (Figure 9F) and Evo + PEI-EVs (Figure 9E), the implant site was covered with an organized extracellular matrix showing mineralization areas and blood-vessel formation. In particular, in harvested Evo + PEI-EVs + hPDLSC samples, valuable osteoblast-like structures were seen on the host-bone side (Figure 9F). A summary of these results is reported in Table 1.
3-D reconstruction of images acquired with CLSM showed spatial interaction of Evo with hPDLSCs, EVs, and PEI-EVs. Calvarium slices with Evo alone are visible in Figure S5A. Calvarium slices in the presence of Evo + EVs and Evo + PEI-EVs showed a link between Evo and secreted PKH76-labeled vesicles at the spatial level (Figure S5B and C, respectively). Moreover, a large number of PKH26-labeled hPDLSCs, easily recognizable by the red lipophilic dye (Figure S5D) and linked to PKH76-stained EVs or PEI-EVs (green), were localized at the cytoplasmic membrane level and on the Evo surface (Figure S5E and F, respectively). The pictures showed good integration of Evo + hPDLSCs, Evo + EVs + hPDLSCs, and Evo + PEI-EVs + hPDLSCs with host tissue.
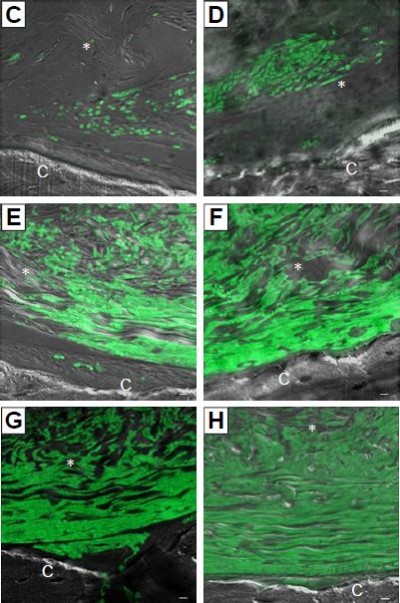
To support the data obtained for gene expression, we focused our attention on the expression of BMP2/4 in in vitro and in vivo conditions. qRT-PCR showed increased expression of BMP2/4 in Evo + PEI-EVs + hPDLSCs when compared with other groups (P<0.01, Figure 10A). Western blotting validated this result (Figure 10B). Moreover, a possible correspondence between upregulated genes in in vitro and in vivo conditions was assayed by means of immunofluorescence analysis of BMP2/4 protein. Data obtained confirmed the in vitro results (Figure 10C–H). In particular, increased expression of positive cells that were round and elongated was visible in the examined samples, indicating a de novo ossification process.
Discussion
Nowadays, bone regeneration represents a clinical challenge. In particular, it is difficult to obtain complete regeneration of calvarium defects caused by trauma and/or other pathologies based only on the intrinsic regenerative capacity of bone. Current treatments include autografts and allografts, but they present some limitations, such as donor-site pain, morbidity, possibility of rejection, disease transmission, and inflammation. In this context, one strategy to overcome defective bone repair and improve bone-tissue regeneration, increasing the rate and quality of the new bone, could be to identify and apply a scaffold-SC construct on bone defects that can inter alia enhance engraftment and paracrine activity of resident SCs. In particular, this approach allows the harvesting of MSCs directly from the patient, and dental SC collection is easy, requiring only minimal invasive procedures. Autologous cell-based approaches are ideal to minimize immunorejection, and today represent a promising new strategy in tissue regeneration after the decline in cellular function associated with aging or in pathological conditions.
MSCs have been applied in autologous therapy in combination with platelet-rich plasma and/or scaffolds in distraction osteogenesis.38,39 Recently, our data confirmed that hPDLSCs seeded onto 3-D dual-block scaffolds promote bone regeneration.10 This effect was due to the ability of SCs to undergo osteogenic differentiation and secrete SDF1, which is able to promote cellular homing and MSC immunomodulatory properties, maximizing regenerative effects with an increase in tissue restoration.10,40 Moreover, the CM and EVs released from hPDLSCs reduced proinflammatory cytokines and induced anti-inflammatory IL10 molecular secretion,13 which are crucial components in bone regeneration. EVs released from eukaryotic cells, like exosomes, microparticles, microvesicles, and apoptotic bodies, can be retained as a dynamic extracellular vesicular compartment, strategic for their paracrine or autocrine biological effects on tissue metabolism.41 In vitro exosomes originating from mineralizing osteoblasts have been demonstrated capability of being able to enter BM stromal cells and inducing osteoblast commitment through the upregulation of β-catenin, thus opening a new tactic in the use of SC-secreted products as a potential therapeutic approach.42
Recently, it has been shown that a human-induced pluripotent SC–MSC–exosome-functionalized β-TCP scaffold effectively promoted bone repair and regeneration in a rat model of calvarium bone defects, thanks to exosomes released from the exosome–β-TCP complex and then internalized by BMSCs enhancing the osteoinductive activity of β-TCP and promoting bone regeneration, due to the activation of endogenous BMSCs in the bone-defect site.43 EVs released from stem/progenitor cells have been shown to possess proregenerative ability in animal models using several signaling pathways involved in EV-mediated tissue regeneration.44
The reconstruction of bone defects is still a challenging problem in orthopedic surgery. Biofunctionalized materials can offer a novel solution to autologous and allogeneic bone grafting in bone repair and regeneration.44 The present study focused on the therapeutic potential of EVs to promote tissue repair, regeneration, and mainly bone regeneration using a commercial collagen membrane (Evo) functionalized with EVs alone or complexed with PEI to stimulate SC viability and activity. Many features of PEI have been shown to affect its efficiency when used as a transfecting agent, eg, molecular weight, degree of branching, ζ-potential, molecular structure, and particle size.45 We found that the best compromise between effectiveness and toxicity was a concentration of 0.05 mg/mL PEI, which conferred to EVs a slight increase in ζ-potential from −10.7±0.9 mV to −1.2±0.9 mV with tolerable viability of recipient cells (5%–10%).
The ability of PEI-EVs to promote internalization with respect to EVs could be ascribed to the positively charged PEI that favors binding to negatively charged proteoglycans.46 Different mechanisms have been proposed for EV internalization: fusion or attachment to the target-cell membrane and subsequent release of exosomal proteins to the recipient cell47,48 or endocytosis.49 In the present study, labeled PEI-EVs were internalized prevailingly by endocytosis, with endosomes highly represented after incubation in recipient cells. Another important advantage of PEI was its proton-sponge effect, which has been demonstrated to be essential for endosomal content release and could also explain the superior efficiency of PEI-EVs compared to nonengineered EVs.50
Alizarin red staining provided evidence after 6 weeks of culture in basal conditions that calcium deposits were frequently observed in the presence of EVs, and, in particular, with PEI-EVs. According to these results, in the presence of PEI-EVs, qRT-PCR showed upregulation of key genes involved in the bone-differentiation pathway, such as TUFT1, TFIP11, BMP2–BMP4, and TGFB. TUFT1 expression in cartilage has demonstrated to be strongest in the deep, mineralizing zones,51 and its expression has been shown to be regulated in vitro by HIF1α and Hedgehog pathways, both essential elements involved in cartilage and bone formation.52 TFIP11 is a gene involved in pre-mRNA splicing, specifically in spliceosome disassembly during late-stage splicing events that may play a role in the differentiation of ameloblasts and odontoblasts or in the formation of the extracellular mineralized matrix.53 Associated with skeletal development and extracellular matrix proteins, TFIP11 is upregulated in mice after tetrachlorodibenzo-p-dioxin exposure.54 Interestingly, TUFT1 and TFIP11 can affect the formation of dental enamel.55
Originally isolated as proteins that induce bone and cartilage formation, BMPs are considered to comprise almost a third of TGFβ. The BMP family is the largest within the TGFβ superfamily of growth factors, which also includes TGFβ, activin, inhibin, myostatin, and others.56 BMPs are potent osteoblast differentiation factors in vitro. In particular BMP2, BMP4, and BMP7 are responsible for the osteochondrogenic differentiation of multipotential mesenchymal cells.57 TGFβ1 promotes matrix production and osteoblast differentiation while reducing the ability of osteoblasts to secrete the osteoclast-differentiation factor RANKL. Thereby, TGFβ1 indirectly limits further osteoclast formation and may affect bone mass.58 Our results highlighted the key role of the TGFβ–BMP pathway in new-bone formation in EV-enriched collagen membrane, and this result was even more evident in PEI-EVs. Previously, we have demonstrated that hPDLSCs implanted in rats did not show immunogenic effects, and after 3 weeks a massive number of cells of different size and features and maturation degree were detected in rat calvaria implanted with hPDLSCs/porcine corticocancellous scaffold (OsteoBiol dual block).10
Here, we demonstrated that hPDLSCs seeded onto Evo were able to induce a bone-regeneration process, but the presence of EVs and mainly PEI-EVs activated local osteogenic induction, hardly contributing to the regeneration process. In fact, Evo scaffold implanted in rat calvaria did not show immunogenic effects, and after 6 weeks numerous cells with fibroblast-like morphology secreting extracellular matrix were observed. On the contrary, the presence of the EVs and more specifically PEI-EVs linked to the scaffold improved the mineralization process and induced an extensive vascular network, indicating an osseointegration process. This condition suggested that the subcellular fraction of EVs directly contributed to the osteogenic regeneration, and in particular PEI-EVs were responsible for more rapid evolution and a major part of maturation of new-bone tissue. These data have been confirmed by the demonstration that PEI-decorated graphene oxide was highly potent in inducing SC osteogenesis, leading to near doubling of alkaline phosphatase expression and mineralization.59
In our experiments, PEI-EVs had a strategic role in determining cell fate, showing that rather than being inert,60 PEI own chemical and physical properties that in combination with EVs, synergistically exert a positive effect on cell morphology and gene transcription by increasing the ability to differentiate along the osteogenic lineage. These data support a new concept of personalized therapy in which EVs and PEI-EVs may be collected by hPDLSCs isolated directly from the patient, avoiding in this way the risk of immunorejection and infections. EVs and PEI-EVs are regarded as a potentially safer and cost-effective alternative for bone-tissue clinical purposes. Moreover, EVs could be applied to specific tissue.
Conclusion
We suggest that Evo enriched with hPDLSCs and PEI-EVs is capable of inducing bone regeneration. In particular, PEI-EVs played a key role in the activation of the osteogenic regenerative process. Indeed, the presence of PEI-EVs improved the mineralization process and induced an extensive vascular network, suggesting an osseointegration process. These data encourage a deep investigation of PEI-EVs, in order to use them in bone-tissue regeneration in combination with different types of scaffolds and SCs.
Acknowledgments
This work was supported by the Oriana Trubiani ex 60% University of Chieti–Pescara Fund, and partly by Progetti di Ricerca di Rilevante Interesse Nazionale grant number 20102ZLNJ5 financed by the Ministry of Education, University, and Research (MIUR), Rome, Italy. This study was also supported by funds of Current Research 2018 of Bonino-Pulejo IRCCS Centro Neurolesi, Messina, Italy. The authors thank Alessia Ventrella, PhD student of the University of Chieti–Pescara, for helping with the engineering of some of the EVs.
Disclosure
The authors report no conflicts of interest in this work.
References
Florencio-Silva R, Sasso GR, Sasso-Cerri E, Simões MJ, Cerri PS. Biology of bone tissue: structure, function, and factors that influence bone cells. Biomed Res Int. 2015;2015:421746. | ||
Dahlin C, Linde A, Gottlow J, Nyman S. Healing of bone defects by guided tissue regeneration. Plast Reconstr Surg. 1988;81(5):672–676. | ||
Basha RY, Kumar TS, Doble M. Design of biocomposite materials for bone tissue regeneration. Mater Sci Eng C Mater Biol Appl. 2015;57:452–463. | ||
Ferreira AM, Gentile P, Chiono V, Ciardelli G. Collagen for bone tissue regeneration. Acta Biomater. 2012;8(9):3191–3200. | ||
Chu C, Deng J, Hou Y, et al. Application of PEG and EGCG modified collagen-base membrane to promote osteoblasts proliferation. Mater Sci Eng C Mater Biol Appl. 2017;76:31–36. | ||
Bojic S, Volarevic V, Ljujic B, Stojkovic M. Dental stem cells: characteristics and potential. Histol Histopathol. 2014;29(6):699–706. | ||
Sedgley CM, Botero TM. Dental stem cells and their sources. Dent Clin North Am. 2012;56(3):549–561. | ||
Rajan TS, Scionti D, Diomede F, et al. Gingival stromal cells as an in vitro model: cannabidiol modulates genes linked with amyotrophic lateral sclerosis. J Cell Biochem. 2017;118(4):819–828. | ||
Achilleos A, Trainor PA. Neural crest stem cells: discovery, properties and potential for therapy. Cell Res. 2012;22(2):288–304. | ||
Diomede F, Zini N, Gatta V, et al. Human periodontal ligament stem cells cultured onto cortico-cancellous scaffold drive bone regenerative process. Eur Cell Mater. 2016;32:181–201. | ||
Zhang C, Yan B, Cui Z, et al. Bone regeneration in minipigs by intrafibrillarly-mineralized collagen loaded with autologous periodontal ligament stem cells. Sci Rep. 2017;7(1):10519. | ||
Cianci E, Recchiuti A, Trubiani O, et al. Human periodontal stem cells release specialized proresolving mediators and carry immunomodulatory and prohealing properties regulated by lipoxins. Stem Cells Transl Med. 2016;5(1):20–32. | ||
Rajan TS, Giacoppo S, Diomede F, et al. The secretome of periodontal ligament stem cells from MS patients protects against EAE. Sci Rep. 2016;6:38743. | ||
Jing H, He X, Zheng J. Exosomes and regenerative medicine: state of the art and perspectives. Transl Res. 2018;196:1–16. | ||
Hao ZC, Lu J, Wang SZ, Wu H, Zhang YT, Xu SG. Stem cell-derived exosomes: a promising strategy for fracture healing. Cell Prolif. 2017;50(5):e12359. | ||
Qin Y, Wang L, Gao Z, Chen G, Zhang C. Bone marrow stromal/stem cell-derived extracellular vesicles regulate osteoblast activity and differentiation in vitro and promote bone regeneration in vivo. Sci Rep. 2016;6:21961. | ||
Furuta T, Miyaki S, Ishitobi H, et al. Mesenchymal stem cell-derived exosomes promote fracture healing in a mouse model. Stem Cells Transl Med. 2016;5(12):1620–1630. | ||
Zhang S, Chu WC, Lai RC, Lim SK, Hui JH, Toh WS. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthritis Cartilage. 2016;24(12):2135–2140. | ||
Boussif O, Lezoualch F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in-vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–7301. | ||
Marshall PL, King JL, Budowle B. Utility of amplification enhancers in low copy number DNA analysis. Int J Legal Med. 2015;129(1):43–52. | ||
Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Hum Gene Ther. 1996;7(16):1947–1954. | ||
Boletta A, Benigni A, Lutz J, Remuzzi G, Soria MR, Monaco L. Nonviral gene delivery to the rat kidney with polyethylenimine. Hum Gene Ther. 1997;8(10):1243–1251. | ||
Goula D, Benoist C, Mantero S, Merlo G, Levi G, Demeneix BA. Polyethylenimine-based intravenous delivery of transgenes to mouse lung. Gene Ther. 1998;5(9):1291–1295. | ||
Werth S, Urban-Klein B, Dai L, et al. A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. J Control Release. 2006;112(2):257–270. | ||
Zuber G, Dauty E, Nothisen M, Belguise P, Behr JP. Towards synthetic viruses. Adv Drug Deliv Rev. 2001;52(3):245–253. | ||
Diomede F, Caputi S, Merciaro I, et al. Pro-inflammatory cytokine release and cell growth inhibition in primary human oral cells after exposure to endodontic sealer. Int Endod J. 2014;47(9):864–872. | ||
Diomede F, Zingariello M, Cavalcanti M, et al. MyD88/ERK/NFκB pathways and pro-inflammatory cytokines release in periodontal ligament stem cells stimulated by Porphyromonas gingivalis. Eur J Histochem. 2017;61(2):2791. | ||
Rajan TS, Giacoppo S, Trubiani O, et al. Conditioned medium of periodontal ligament mesenchymal stem cells exert anti-inflammatory effects in lipopolysaccharide-activated mouse motoneurons. Exp Cell Res. 2016;349(1):152–161. | ||
Ballerini P, Diomede F, Petragnani N, et al. Conditioned medium from relapsing-remitting multiple sclerosis patients reduces the expression and release of inflammatory cytokines induced by LPS-gingivalis in THP-1 and MO3.13 cell lines. Cytokine. 2017;96:261–272. | ||
Angelini G, Boncompagni S, De Maria P, et al. Layer-by-layer deposition of shortened nanotubes or polyethylene glycol-derivatized nanotubes on liposomes: a tool for increasing liposome stability. Carbon. 2007;45(13):2479–2485. | ||
Angelini G, Boncompagni S, De Maria P, Fontana A, Gasbarri C, Siani G. Kinetic evaluation of the effect of layer by layer deposition of polyelectrolytes on the stability of POPC liposomes. Colloids Surf A Physicochem Eng Asp. 2008;322(1–3):234–238. | ||
Diomede F, Rajan TS, Gatta V, et al. Stemness maintenance properties in human oral stem cells after long-term passage. Stem Cells Int. 2017;2017:5651287. | ||
Orciani M, Trubiani O, Guarnieri S, Ferrero E, Di Primio R. CD38 is constitutively expressed in the nucleus of human hematopoietic cells. J Cell Biochem. 2008;105(3):905–912. | ||
Trubiani O, Guarnieri S, Diomede F, et al. Nuclear translocation of PKCα isoenzyme is involved in neurogenic commitment of human neural crest-derived periodontal ligament stem cells. Cell Signal. 2016;28(11):1631–1641. | ||
Libro R, Scionti D, Diomede F, et al. Cannabidiol modulates the immunophenotype and inhibits the activation of the inflammasome in human gingival mesenchymal stem cells. Front Physiol. 2016;7:559. | ||
Gugliandolo A, Diomede F, Cardelli P, et al. Transcriptomic analysis of gingival mesenchymal stem cells cultured on 3D bioprinted scaffold: a promising strategy for neuroregeneration. J Biomed Mater Res A. 2018;106(1):126–137. | ||
Sharma S, Rasool HI, Palanisamy V, et al. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010;4(4):1921–1926. | ||
Yamada Y, Ueda M, Naiki T, Takahashi M, Hata K, Nagasaka T. Autogenous injectable bone for regeneration with mesenchymal stem cells and platelet-rich plasma: tissue-engineered bone regeneration. Tissue Eng. 2004;10(5–6):955–964. | ||
Yamada Y, Ueda M, Hibi H, Nagasaka T. Translational research for injectable tissue-engineered bone regeneration using mesenchymal stem cells and platelet-rich plasma: from basic research to clinical case study. Cell Transplant. 2004;13(4):343–355. | ||
Mountziaris PM, Spicer PP, Kasper FK, Mikos AG. Harnessing and modulating inflammation in strategies for bone regeneration. Tissue Eng Part B Rev. 2011;17(6):393–402. | ||
Gyorgy B, Szabo TG, Pasztoi M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667–2688. | ||
Cui Y, Luan J, Li H, Zhou X, Han J. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS Lett. 2016;590(1):185–192. | ||
Zhang JY, Liu XL, Li HY, et al. Exosomes/tricalcium phosphate combination scaffolds can enhance bone regeneration by activating the PI3K/Akt signaling pathway. Stem Cell Res Ther. 2016;7(1):136. | ||
Chen B, Li Q, Zhao BZ, Wang Y. Stem cell-derived extracellular vesicles as a novel potential therapeutic tool for tissue repair. Stem Cells Transl Med. 2017;6(9):1753–1758. | ||
Werth S, Urban-Klein B, Dai L, et al. A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. J Control Release. 2006;112(2):257–270. | ||
Mislick KA, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci U S A. 1996;93(22):12349–12354. | ||
Denzer K, van Eijk M, Kleijmeer MJ, Jakobson E, de Groot C, Geuze HJ. Follicular dendritic cells carry MHC class II-expressing microvesicles at their surface. J Immunol. 2000;165(3):1259–1265. | ||
Clayton A, Turkes A, Dewitt S, Steadman R, Mason MD, Hallett MB. Adhesion and signaling by B cell-derived exosomes: the role of integrins. FASEB J. 2004;18(6):977–979. | ||
Morelli AE, Larregina AT, Shufesky WJ, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104(10):3257–3266. | ||
Silva AM, Almeida MI, Teixeira JH, et al. Dendritic cell-derived extracellular vesicles mediate mesenchymal stem/stromal cell recruitment. Sci Rep. 2017;7(1):1667. | ||
Sliz E, Taipale M, Welling M, et al. TUFT1, a novel candidate gene for metatarsophalangeal osteoarthritis, plays a role in chondrogenesis on a calcium-related pathway. PLoS One. 2017;12(4):e0175474. | ||
Rankin EB, Giaccia AJ, Schipani E. A central role for hypoxic signaling in cartilage, bone, and hematopoiesis. Curr Osteoporos Rep. 2011;9(2):46–52. | ||
Yoshimoto R, Kataoka N, Okawa K, Ohno M. Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res. 2009;37(3):891–902. | ||
Herlin M, Finnila MA, Zioupos P, et al. New insights to the role of aryl hydrocarbon receptor in bone phenotype and in dioxin-induced modulation of bone microarchitecture and material properties. Toxicol Appl Pharmacol. 2013;273(1):219–226. | ||
Gerreth K, Zaorska K, Zabel M, Nowicki M, Borysewicz-Lewicka M. Significance of genetic variations in developmental enamel defects of primary dentition in Polish children. Clin Oral Investig. 2018;22(1):321–329. | ||
Shi Y, Massague J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. | ||
Balint E, Lapointe D, Drissi H, et al. Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J Cell Biochem. 2003;89(2):401–426. | ||
Yu J, Xu L, Li K, et al. Zinc-modified calcium silicate coatings promote osteogenic differentiation through TGF-β/Smad pathway and osseointegration in osteopenic rabbits. Sci Rep. 2017;7(1):3440. | ||
Kumar S, Raj S, Sarkar K, Chatterjee K. Engineering a multi-biofunctional composite using poly(ethylenimine) decorated graphene oxide for bone tissue regeneration. Nanoscale. 2016;8(12):6820–6836. | ||
Gonzalez-Fernandez T, Sathy BN, Hobbs C, et al. Mesenchymal stem cell fate following non-viral gene transfection strongly depends on the choice of delivery vector. Acta Biomater. 2017;55:226–238. |
Supplementary materials
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.